1QN6
 
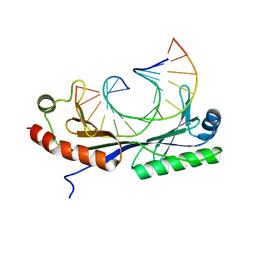 | | Crystal structure of the T(-26) Adenovirus major late promoter TATA box variant bound to wild-type TBP (Arabidopsis thaliana TBP isoform 2). TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*TP*AP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*TP*AP*TP*TP*AP*TP*AP*GP*C)-3'), TRANSCRIPTION INITIATION FACTOR TFIID-1 | | Authors: | Patikoglou, G.A, Kim, J.L, Sun, L, Yang, S.-H, Kodadek, T, Burley, S.K. | | Deposit date: | 1999-10-14 | | Release date: | 2000-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | TATA Element Recognition by the TATA Box-Binding Protein Has Been Conserved Throughout Evolution
Genes Dev., 13, 1999
|
|
1QN8
 
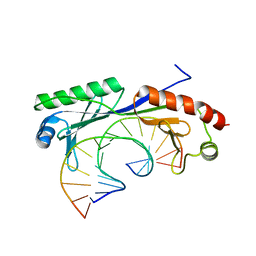 | | Crystal structure of the T(-28) Adenovirus major late promoter TATA box variant bound to wild-type TBP (Arabidopsis thaliana TBP isoform 2). TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*TP*TP*AP*AP*AP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*TP*TP*TP*AP*AP*TP*AP*GP*C)-3'), TRANSCRIPTION INITIATION FACTOR TFIID-1 | | Authors: | Patikoglou, G.A, Kim, J.L, Sun, L, Yang, S.-H, Kodadek, T, Burley, S.K. | | Deposit date: | 1999-10-14 | | Release date: | 2000-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | TATA Element Recognition by the TATA Box-Binding Protein Has Been Conserved Throughout Evolution
Genes Dev., 13, 1999
|
|
1QNA
 
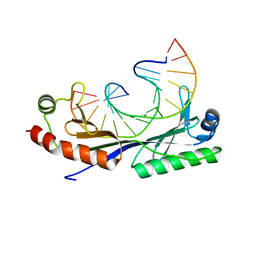 | | Crystal structure of the T(-30) Adenovirus major late promoter TATA box variant bound to wild-type TBP (Arabidopsis thaliana TBP isoform 2). TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. | | Descriptor: | DNA (5'-D(*GP*CP*TP*TP*TP*AP*AP*AP*AP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*TP*TP*TP*TP*AP*AP*AP*GP*C)-3'), TRANSCRIPTION INITIATION FACTOR TFIID-1 | | Authors: | Patikoglou, G.A, Kim, J.L, Sun, L, Yang, S.-H, Kodadek, T, Burley, S.K. | | Deposit date: | 1999-10-14 | | Release date: | 2000-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | TATA Element Recognition by the TATA Box-Binding Protein Has Been Conserved Throughout Evolution
Genes Dev., 13, 1999
|
|
1QN3
 
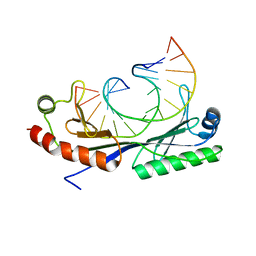 | | Crystal structure of the C(-25) Adenovirus major late promoter TATA box variant bound to wild-type TBP (Arabidopsis thaliana TBP isoform 2). TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*AP*CP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*GP*TP*TP*TP*AP*TP*AP*GP*C)-3'), TRANSCRIPTION INITIATION FACTOR TFIID-1 | | Authors: | Patikoglou, G.A, Kim, J.L, Sun, L, Yang, S.-H, Kodadek, T, Burley, S.K. | | Deposit date: | 1999-10-14 | | Release date: | 2000-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | TATA Element Recognition by the TATA Box-Binding Protein Has Been Conserved Throughout Evolution
Genes Dev., 13, 1999
|
|
1QN7
 
 | | Crystal structure of the T(-27) Adenovirus major late promoter TATA box variant bound to wild-type TBP (Arabidopsis thaliana TBP isoform 2). TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*TP*AP*AP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*TP*TP*AP*TP*AP*TP*AP*GP*C)-3'), TRANSCRIPTION INITIATION FACTOR TFIID-1 | | Authors: | Patikoglou, G.A, Kim, J.L, Sun, L, Yang, S.-H, Kodadek, T, Burley, S.K. | | Deposit date: | 1999-10-14 | | Release date: | 2000-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | TATA Element Recognition by the TATA Box-Binding Protein Has Been Conserved Throughout Evolution
Genes Dev., 13, 1999
|
|
1QN5
 
 | | Crystal structure of the G(-26) Adenovirus major late promoter TATA box variant bound to wild-type TBP (Arabidopsis thaliana TBP isoform 2). TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*GP*AP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*TP*CP*TP*TP*AP*TP*AP*GP*C)-3'), TRANSCRIPTION INITIATION FACTOR TFIID-1 | | Authors: | Patikoglou, G.A, Kim, J.L, Sun, L, Yang, S.-H, Kodadek, T, Burley, S.K. | | Deposit date: | 1999-10-14 | | Release date: | 2000-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | TATA Element Recognition by the TATA Box-Binding Protein Has Been Conserved Throughout Evolution
Genes Dev., 13, 1999
|
|
1RIO
 
 | | Structure of bacteriophage lambda cI-NTD in complex with sigma-region4 of Thermus aquaticus bound to DNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 27-MER, CALCIUM ION, ... | | Authors: | Jain, D, Nickels, B.E, Sun, L, Hochschild, A, Darst, S.A. | | Deposit date: | 2003-11-17 | | Release date: | 2004-01-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a ternary transcription activation complex.
Mol.Cell, 13, 2004
|
|
8I4F
 
 | | Omicron spike variant XBB with n3130v-Fc | | Descriptor: | Spike glycoprotein, n3130v-Fc | | Authors: | Hao, A.H, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2023-01-19 | | Release date: | 2023-12-27 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants.
Signal Transduct Target Ther, 8, 2023
|
|
8I4G
 
 | | Omicron spike variant BQ.1.1 with n3130v-Fc | | Descriptor: | Spike glycoprotein, n3130v-Fc | | Authors: | Hao, A.H, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2023-01-19 | | Release date: | 2023-12-27 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants.
Signal Transduct Target Ther, 8, 2023
|
|
8I4E
 
 | | Omicron spike variant XBB with Bn03 | | Descriptor: | Bn03, Spike glycoprotein | | Authors: | Hao, A.H, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2023-01-19 | | Release date: | 2023-12-27 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants.
Signal Transduct Target Ther, 8, 2023
|
|
8I4H
 
 | | Omicron spike variant BA.1 with Bn03 | | Descriptor: | Bn03, Spike glycoprotein | | Authors: | Hao, A.H, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-31 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants.
Signal Transduct Target Ther, 8, 2023
|
|
8HZN
 
 | |
2IS9
 
 | | Structure of yeast DCN-1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Defective in cullin neddylation protein 1, ... | | Authors: | Yang, X, Zhou, J, Sun, L, Wei, Z, Gao, J, Gong, W, Xu, R.M, Rao, Z, Liu, Y. | | Deposit date: | 2006-10-16 | | Release date: | 2007-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for the function of DCN-1 in protein Neddylation.
J.Biol.Chem., 282, 2007
|
|
6CO8
 
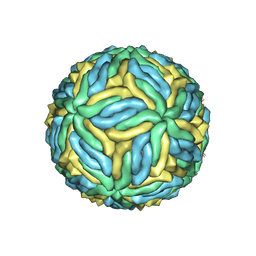 | | Structure of Zika virus at a resolution of 3.1 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, E protein, M protein | | Authors: | Sevvana, M, Long, F, Miller, A.J, Klose, T, Buda, G, Sun, L, Kuhn, R.J, Rossmann, M.R. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Refinement and Analysis of the Mature Zika Virus Cryo-EM Structure at 3.1 angstrom Resolution.
Structure, 26, 2018
|
|
8WOO
 
 | | Structure of the wild-type Arabidopsis ABCB19 in the brassinolide and AMP-PNP bound state | | Descriptor: | ABC transporter B family member 19, Brassinolide, MAGNESIUM ION, ... | | Authors: | Ying, W, Wei, H, Liu, X, Sun, L. | | Deposit date: | 2023-10-07 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure and function of the Arabidopsis ABC transporter ABCB19 in brassinosteroid export.
Science, 383, 2024
|
|
8WOM
 
 | | Structure of the wild-type Arabidopsis ABCB19 in the brassinolide-bound state | | Descriptor: | ABC transporter B family member 19, Brassinolide | | Authors: | Ying, W, Wei, H, Liu, X, Sun, L. | | Deposit date: | 2023-10-07 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and function of the Arabidopsis ABC transporter ABCB19 in brassinosteroid export.
Science, 383, 2024
|
|
8WOI
 
 | |
8WP0
 
 | | Structure of the Arabidopsis E529Q/E1174Q ABCB19 in the ATP bound state | | Descriptor: | ABC transporter B family member 19, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Ying, W, Wei, H, Liu, X, Sun, L. | | Deposit date: | 2023-10-08 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and function of the Arabidopsis ABC transporter ABCB19 in brassinosteroid export.
Science, 383, 2024
|
|
8HEI
 
 | | Crystal structure of CTSB in complex with E64d | | Descriptor: | Cathepsin B, GLYCEROL, ethyl (3S)-3-hydroxy-4-({(2S)-4-methyl-1-[(3-methylbutyl)amino]-1-oxopentan-2-yl}amino)-4-oxobutanoate | | Authors: | Wang, H, Li, D, Sun, L, Yang, H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HE9
 
 | | Crystal structure of CTSB in complex with K777 | | Descriptor: | Cathepsin B, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Wang, H, Li, D, Sun, L, Yang, H. | | Deposit date: | 2022-11-07 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HFV
 
 | | Crystal structure of CTSL in complex with K777 | | Descriptor: | CACODYLATE ION, Nalpha-[(4-methylpiperazin-1-yl)carbonyl]-N-[(3S)-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]-L-phenylalaninamide, Procathepsin L, ... | | Authors: | Wang, H, Shao, M, Sun, L, Yang, H. | | Deposit date: | 2022-11-12 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HET
 
 | | Crystal structure of CTSL in complex with E64d | | Descriptor: | Procathepsin L, ethyl (3S)-3-hydroxy-4-({(2S)-4-methyl-1-[(3-methylbutyl)amino]-1-oxopentan-2-yl}amino)-4-oxobutanoate | | Authors: | Wang, H, Shao, M, Sun, L, Yang, H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HD8
 
 | | Crystal structure of TMPRSS2 in complex with 212-148 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, CALCIUM ION, ... | | Authors: | Wang, H, Liu, X, Sun, L, Yang, H. | | Deposit date: | 2022-11-03 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HEN
 
 | | Crystal structure of CTSB in complex with 212-148 | | Descriptor: | 2-[4-[[(2~{S})-1-oxidanylidene-3-phenyl-1-[[(3~{S})-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]amino]propan-2-yl]carbamoyl]piperazin-1-yl]ethyl 4-carbamimidamidobenzoate, Cathepsin B, DIMETHYL SULFOXIDE, ... | | Authors: | Wang, H, Li, D, Sun, L, Yang, H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
7XYD
 
 | | Crystal structure of TMPRSS2 in complex with Nafamostat | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, CALCIUM ION, ... | | Authors: | Wang, H, Liu, X, Duan, Y, Liu, X, Sun, L, Yang, H. | | Deposit date: | 2022-06-01 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
