3N1D
 
 | | Crystal structure of the complex of type I ribosome inactivating protein with ribose at 1.7A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Pandey, N, Kushwaha, G.S, Sinha, M, Kaur, P, Betzel, C, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-15 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the complex of type I ribosome inactivating protein with ribose at 1.7A resolution
To be Published
|
|
3MU7
 
 | | Crystal structure of the xylanase and alpha-amylase inhibitor protein (XAIP-II) from scadoxus multiflorus at 1.2 A resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, xylanase and alpha-amylase inhibitor protein | | Authors: | Kumar, S, Singh, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-02 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Modulation of inhibitory activity of xylanase-alpha-amylase inhibitor protein (XAIP): binding studies and crystal structure determination of XAIP-II from Scadoxus multiflorus at 1.2 A resolution.
Bmc Struct.Biol., 10, 2010
|
|
3N8F
 
 | | Crystal structure of the complex of goat lactoperoxidase with thiocyanate at 3.2 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Lactoperoxidase, ... | | Authors: | Vikram, G, Singh, A.K, Singh, R.P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-28 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of the complex of goat lactoperoxidase with thiocyanate at 3.2 A resolution
To be Published
|
|
3N2D
 
 | | Crystal Structure of the Complex of type I Ribosome inactivating protein with hexapeptide Ser-Asp-Asp-Asp-Met-Gly at 2.2 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-meric peptide from 60S acidic ribosomal protein P2-beta, Ribosome inactivating protein | | Authors: | Kushwaha, G.S, Prem Kumar, R, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-18 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal Structure of the Complex of type I Ribosome inactivating protein with hexapeptide Ser-Asp-Asp-Asp-Met-Gly at 2.2 A resolution
To be Published
|
|
3N5D
 
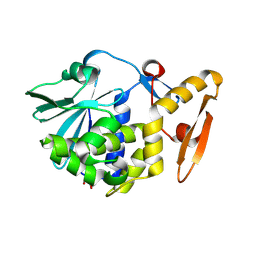 | | Crystal structure of the complex of type I ribosome inactivating protein with glucose at 1.9A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Pandey, N, Kushwaha, G.S, Sinha, M, Kaur, P, Betzel, C, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-25 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the complex of type I ribosome inactivating protein with glucose at 1.9A resolution
To be Published
|
|
3N1N
 
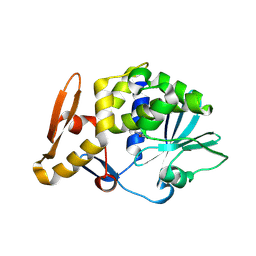 | | Crystal structure of the complex of type I ribosome inactivating protein with guanine at 2.2A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GUANINE, Ribosome inactivating protein | | Authors: | Kushwaha, G.S, Singh, N, Sinha, M, Kaur, P, Betzel, C, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-16 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structures of a type-1 ribosome inactivating protein from Momordica balsamina in the bound and unbound states
Biochim.Biophys.Acta, 1824, 2012
|
|
3NIU
 
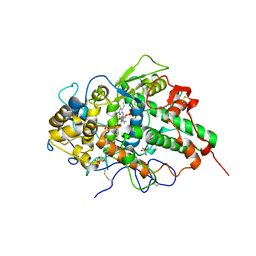 | | Crystal structure of the complex of dimeric goat lactoperoxidase with diethylene glycol at 2.9 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Vikram, G, Singh, R.P, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-16 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Crystal structure of the complex of dimeric goat lactoperoxidase with diethylene glycol at 2.9 A resolution
To be Published
|
|
3N3X
 
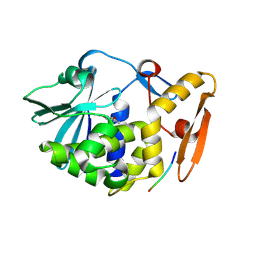 | | Crystal Structure of the complex formed between type I ribosome inactivating protein and hexapeptide Ser-Asp-Asp-Asp-Met-Gly at 1.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GUANINE, Ribosome inactivating protein, ... | | Authors: | Kushwaha, G.S, Vikram, G, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the complex formed between type I ribosome inactivating protein and hexapeptide Ser-Asp-Asp-Asp-Met-Gly at 1.7 A resolution
To be Published
|
|
3NFM
 
 | | Crystal Structure of the complex of type I ribosome inactivating protein with fructose at 2.5A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Kushwaha, G.S, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-10 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the complex of type I ribosome inactivating protein with fructose at 2.5A resolution
To be Published
|
|
3NJS
 
 | | Crystal structure of the complex formed between typeI ribosome inactivating protein and lactose at 2.1A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Pandey, N, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the complex formed between typeI ribosome inactivating protein and lactose at 2.1A resolution
To be Published
|
|
4EMR
 
 | | Crystal Structure determination of type1 ribosome inactivating protein complexed with 7-methylguanosine-triphosphate at 1.75A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, rRNA N-glycosidase | | Authors: | Kumar, M, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | First structural evidence of sequestration of mRNA cap structures by type 1 ribosome inactivating protein from Momordica balsamina.
Proteins, 81, 2013
|
|
4DXV
 
 | | Crystal structure of Dihydrodipicolinate synthase from Acinetobacter baumannii complexed with Mg and Cl ions at 1.80 A resolution | | Descriptor: | CHLORIDE ION, Dihydrodipicolinate synthase, GLYCEROL, ... | | Authors: | Kumar, M, Kaushik, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-02-28 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Dihydrodipicolinate synthase from Acinetobacter baumannii complexed with Mg and Cl ions at 1.80 A resolution
To be Published
|
|
4GLB
 
 | | Structure of p-nitrobenzaldehyde inhibited lipase from Thermomyces lanuginosa at 2.69 A resolution | | Descriptor: | 4-nitrobenzaldehyde, GLYCEROL, Lipase | | Authors: | Kumar, M, Sinha, M, Mukherjee, J, Gupta, M.N, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-08-14 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure of p-nitrobenzaldehyde inhibited lipase from Thermomyces lanuginosa at 2.69 A resolution
TO BE PUBLISHED
|
|
4GI1
 
 | | Structure of the complex of three phase partition treated lipase from Thermomyces lanuginosa with 16-hydroxypalmitic acid at 2.4 A resolution | | Descriptor: | 16-hydroxyhexadecanoic acid, GLYCEROL, Lipase | | Authors: | Kumar, M, Sinha, M, Mukherjee, J, Gupta, M.N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-08-08 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure of the complex of three phase partition treated lipase from Thermomyces lanuginosa with 16-hydroxypalmitic acid at 2.4 A resolution
To be published
|
|
4FLF
 
 | | Structure of three phase partition treated lipase from Thermomyces lanuginosa at 2.15A resolution. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-nitrobenzaldehyde, ... | | Authors: | Kumar, M, Mukherjee, J, Sinha, M, Kaur, P, Gupta, M.N, Sharma, S, Singh, T.P. | | Deposit date: | 2012-06-14 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Enhancement of stability of a lipase by subjecting to three phase partitioning (TPP): structures of native and TPP-treated lipase from Thermomyces lanuginosa
Sustain Chem Process, 2015
|
|
4EA6
 
 | | Crystal structure of Fungal lipase from Thermomyces(Humicola) lanuginosa at 2.30 Angstrom resolution. | | Descriptor: | Lipase | | Authors: | Kumar, M, Mukherjee, J, Gupta, M.N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-03-22 | | Release date: | 2012-04-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Fungal lipase from Thermomyces(Humicola) lanuginosa at 2.30 Angstrom resolution.
To be Published
|
|
4HMB
 
 | | Crystal Structure of the complex of group II phospholipase A2 with a 3-{3-[(Dimethylamino)methyl]-1H-indol-7-yl}propan-1-ol at 2.21 A Resolution | | Descriptor: | 3-{3-[(DIMETHYLAMINO)METHYL]-1H-INDOL-7-YL}PROPAN-1-OL, Phospholipase A2 VRV-PL-VIIIa | | Authors: | Shukla, P.K, Haridas, M, Chandra, D.N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-10-18 | | Release date: | 2012-11-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal Structure of the complex of group II phospholipase A2 with a 3-{3-[(Dimethylamino)methyl]-1H-indol-7-yl}propan-1-ol at 2.21 A Resolution
To be Published
|
|
3S8H
 
 | | Structure of dihydrodipicolinate synthase complexed with 3-Hydroxypropanoic acid(HPA)at 2.70 A resolution | | Descriptor: | 3-HYDROXY-PROPANOIC ACID, Dihydrodipicolinate synthase | | Authors: | Kumar, M, Kaur, N, Kumar, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-05-28 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of dihydrodipicolinate synthase complexed with 3-Hydroxypropanoic acid(HPA)at 2.70 A resolution
TO BE PUBLISHED
|
|
4EMF
 
 | | Crystal structure of the complex of type I Ribosome inactivating protein in complex with 7n-methyl-8-hydroguanosine-5-p-diphosphate at 1.77 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Yamini, S, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | First structural evidence of sequestration of mRNA cap structures by type 1 ribosome inactivating protein from Momordica balsamina.
Proteins, 81, 2013
|
|
3U8G
 
 | | Crystal structure of the complex of Dihydrodipicolinate synthase from Acinetobacter baumannii with Oxalic acid at 1.80 A resolution | | Descriptor: | Dihydrodipicolinate synthase, OXALATE ION | | Authors: | Kumar, M, Kaushik, S, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-10-17 | | Release date: | 2011-11-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the complex of Dihydrodipicolinate synthase from Acinetobacter baumannii with Oxalic acid at 1.80 A resolution
To be Published
|
|
3TDF
 
 | | Crystal structure of the complex of Dihydrodipicolinate synthase from Acinetobacter baumannii with 2-Ketobutanoic acid at 1.99 A resolution | | Descriptor: | 2-KETOBUTYRIC ACID, Dihydrodipicolinate synthase | | Authors: | Kumar, M, Kaushik, S, Sinha, M, Kaur, P, Tewari, R, Sharma, S, Singh, T.P. | | Deposit date: | 2011-08-11 | | Release date: | 2011-08-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of the complex of Dihydrodipicolinate synthase from Acinetobacter baumannii with 2-Ketobutanoic acid at 1.99 A resolution
To be Published
|
|
4I47
 
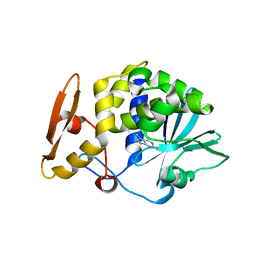 | | Crystal structure of the Ribosome inactivating protein complexed with methylated guanine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-7-methyl-1,7-dihydro-6H-purin-6-one, rRNA N-glycosidase | | Authors: | Yamini, S, Kushwaha, G.S, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-11-27 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | First structural evidence of sequestration of mRNA cap structures by type 1 ribosome inactivating protein from Momordica balsamina.
Proteins, 81, 2013
|
|
3NW3
 
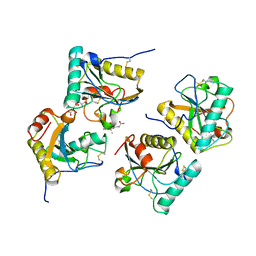 | | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with the PGN Fragment at 2.5 A resolution | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, ALANINE, D-GLUTAMINE, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-07-09 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multiligand specificity of pathogen-associated molecular pattern-binding site in peptidoglycan recognition protein
J.Biol.Chem., 286, 2011
|
|
4N8S
 
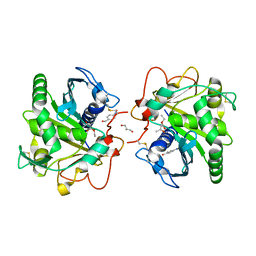 | | Crystal Structure of the ternary complex of lipase from Thermomyces lanuginosa with Ethylacetoacetate and P-nitrobenzaldehyde at 2.3 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-nitrobenzaldehyde, GLYCEROL, ... | | Authors: | Kumar, M, Mukherjee, J, Gupta, M.N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-10-18 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the ternary complex of lipase from Thermomyces lanuginosa with Ethylacetoacetate and P-nitrobenzaldehyde at 2.3 A resolution
To be Published
|
|
4OEK
 
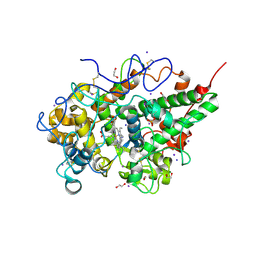 | | Crystal Structure of the Complex of goat Lactoperoxidase with Phenylethylamine at 2.47 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-PHENYLETHYLAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kumar, M, Singh, R.P, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-01-13 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal Structure of the Complex of goat Lactoperoxidase with Phenylethylamine at 2.47 A
To be Published
|
|
