8HSF
 
 | |
8HSK
 
 | |
4DKY
 
 | | Crystal structure Analysis of N terminal region containing the dimerization domain and DNA binding domain of HU protein(Histone like protein-DNA binding) from Mycobacterium tuberculosis [H37Ra] | | Descriptor: | DNA-binding protein HU homolog, MANGANESE (II) ION | | Authors: | Bhowmick, T, Ramagopal, U.A, Ghosh, S, Nagaraja, V, Ramakumar, S. | | Deposit date: | 2012-02-05 | | Release date: | 2013-02-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.478 Å) | | Cite: | Targeting Mycobacterium tuberculosis nucleoid-associated protein HU with structure-based inhibitors
Nat Commun, 5, 2014
|
|
4PT4
 
 | | Crystal structure Analysis of N terminal region containing the dimerization domain and DNA binding domain of HU protein(Histone like protein-DNA binding) from Mycobacterium tuberculosis [H37Ra] | | Descriptor: | DNA-binding protein HU homolog, FORMIC ACID | | Authors: | Bhowmick, T, Ramagopal, U.A, Ghosh, S, Nagaraja, V, Ramakumar, S. | | Deposit date: | 2014-03-10 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Targeting Mycobacterium tuberculosis nucleoid-associated protein HU with structure-based inhibitors
Nat Commun, 5, 2014
|
|
4JHM
 
 | | Crystal structure of a putative mandelate racemase/muconate lactonizing enzyme from Pseudovibrio sp. | | Descriptor: | Mandelate racemase / muconate lactonizing enzyme, C-terminal domain protein | | Authors: | Hegde, R.P, Toro, R, Burley, S.K, Almo, S.C, Ramagopal, U.A, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-05 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a putative mandelate racemase/muconate lactonizing enzyme from Pseudovibrio sp.
To be published
|
|
4MB6
 
 | | Crystal structure of adenine phosphoribosyltransferase from Yersinia pseudotuberculosis. | | Descriptor: | Adenine phosphoribosyltransferase, SODIUM ION | | Authors: | Pavithra, G.C, Kim, J, Hegde, R.P, Almo, S.C, Ramagopal, U.A, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-19 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of adenine phosphoribosyltransferase from Yersinia pseudotuberculosis
To be published
|
|
5KZK
 
 | | Crystal Structure of rRNA methyltransferase from Sinorhizobium meliloti | | Descriptor: | COBALT (II) ION, Probable RNA methyltransferase, TrmH family, ... | | Authors: | Dey, D, Hegde, R.P, Almo, S.C, Ramakumar, S, Ramagopal, U.A, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-07-25 | | Release date: | 2017-08-02 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal Structure of rRNA methyltransferase from Sinorhizobium meliloti
To Be Published
|
|
5L0Z
 
 | | Crystal Structure of AdoMet bound rRNA methyltransferase from Sinorhizobium meliloti | | Descriptor: | COBALT (II) ION, Probable RNA methyltransferase, TrmH family, ... | | Authors: | Dey, D, Hegde, R.P, Almo, S.C, Ramakumar, S, Ramagopal, U.A. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of AdoMet bound rRNA methyltransferase from Sinorhizobium meliloti
To Be Published
|
|
1Q2Y
 
 | | Crystal structure of the protein YJCF from Bacillus subtilis: a member of the GCN5-related N-acetyltransferase superfamily fold | | Descriptor: | similar to hypothetical proteins | | Authors: | Fedorov, A.A, Ramagopal, U.A, Fedorov, E.V, Thirumuruhan, R, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-07-27 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the protein YJCF from Bacillus subtilis: a member of the GCN5-related N-acetyltransferase superfamily
To be Published
|
|
6IGS
 
 | |
6KQC
 
 | |
6KP5
 
 | |
3BGS
 
 | | Structure of human purine nucleoside phosphorylase with L-DADMe-ImmH and phosphate | | Descriptor: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, purine nucleoside phosphorylase | | Authors: | Murkin, A.S, Ramagopal, U.A, Almo, S.C, Schramm, V.L. | | Deposit date: | 2007-11-27 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | L-Enantiomers of transition state analogue inhibitors bound to human purine nucleoside phosphorylase
J.Am.Chem.Soc., 130, 2008
|
|
3BP6
 
 | | Crystal structure of the mouse PD-1 Mutant and PD-L2 complex | | Descriptor: | GLYCEROL, Programmed cell death 1 ligand 2, Programmed cell death protein 1 | | Authors: | Yan, Q, Lazar-Molnar, E, Cao, E, Ramagopal, U.A, Toro, R, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-12-18 | | Release date: | 2009-02-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the mouse PD-1 A99L and PD-L2 complex
To be published
|
|
3B9I
 
 | |
1VRZ
 
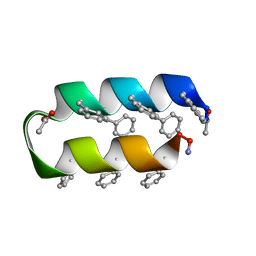 | | Helix turn helix motif | | Descriptor: | ACETATE ION, DE NOVO DESIGNED 21 RESIDUE PEPTIDE | | Authors: | Rudresh, Ramakumar, S, Ramagopal, U.A, Inai, Y, Sahal, D. | | Deposit date: | 2005-10-14 | | Release date: | 2005-11-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | De Novo Design and Characterization of a Helical Hairpin Eicosapeptide; Emergence of an Anion Receptor in the Linker Region.
Structure, 12, 2004
|
|
1XM5
 
 | | Crystal structure of metal-dependent hydrolase ybeY from E. coli, Pfam UPF0054 | | Descriptor: | Hypothetical UPF0054 protein ybeY, NICKEL (II) ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Shi, W, Ramagopal, U.A, Thirumuruhan, R, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-10-01 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The ybeY protein from Escherichia coli is a metalloprotein.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1YK9
 
 | | Crystal structure of a mutant form of the mycobacterial adenylyl cyclase Rv1625c | | Descriptor: | Adenylate cyclase | | Authors: | Ketkar, A.D, Shenoy, A.R, Ramagopal, U.A, Visweswariah, S.S, Suguna, K, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-01-17 | | Release date: | 2006-01-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Structural Basis for the Role of Nucleotide Specifying Residues in Regulating the Oligomerization of the Rv1625c Adenylyl Cyclase from M.tuberculosis
J.Mol.Biol., 356, 2006
|
|
4UC0
 
 | | Crystal Structure Of a purine nucleoside phosphorylase (PSI-NYSGRC-029736) from Agrobacterium vitis | | Descriptor: | HYPOXANTHINE, Purine nucleoside phosphorylase | | Authors: | Cameron, S.A, Sampathkumar, P, Ramagopal, U.A, Attonito, J, Ahmed, M, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Glenn, A.S, Hammonds, J, Hillerich, B, Love, J.D, Seidel, R, Stead, M, Toro, R, Wasserman, S.R, Schramm, V.L, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-08-13 | | Release date: | 2014-10-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure Of a purine nucleoside phosphorylase (PSI-NYSGRC-029736) from Agrobacterium vitis
To be published
|
|
1RC6
 
 | | Crystal structure of protein Ylba from E. coli, Pfam DUF861 | | Descriptor: | Hypothetical protein ylbA | | Authors: | Fedorov, A.A, Fedorov, E.V, Thirumuruhan, R, Ramagopal, U.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-11-03 | | Release date: | 2003-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Ylba, hypothetical protein from E.Coli
To be Published
|
|
2Q3N
 
 | | Agglutinin from Abrus Precatorius (APA-I) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Agglutinin-1 A chain, Agglutinin-1 B chain | | Authors: | Bagaria, A, Surendranath, K, Ramagopal, U.A, Ramakumar, S, Karande, A.A. | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure-Function Analysis and Insights into the Reduced Toxicity of Abrus precatorius Agglutinin I in Relation to Abrin.
J.Biol.Chem., 281, 2006
|
|
2Q91
 
 | | Structure of the Ca2+-Bound Activated Form of the S100A4 Metastasis Factor | | Descriptor: | CALCIUM ION, S100A4 Metastasis Factor | | Authors: | Malashkevich, V.N, Knight, D, Ramagopal, U.A, Almo, S.C, Bresnick, A.R. | | Deposit date: | 2007-06-12 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure of Ca(2+)-Bound S100A4 and Its Interaction with Peptides Derived from Nonmuscle Myosin-IIA.
Biochemistry, 47, 2008
|
|
2QDN
 
 | |
5XTK
 
 | |
5Y4A
 
 | |
