7JJF
 
 | | Sarcin-ricin loop with modified residue. | | Descriptor: | MAGNESIUM ION, RNA/DNA (27-mer) | | Authors: | Harp, J.M, Pallan, P.S, Egli, M. | | Deposit date: | 2020-07-25 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Incorporating a Thiophosphate Modification into a Common RNA Tetraloop Motif Causes an Unanticipated Stability Boost.
Biochemistry, 59, 2020
|
|
7JJD
 
 | |
469D
 
 | | CRYSTAL STRUCTURE AND IMPROVED ANTISENSE PROPERTIES OF 2'-O-(2-METHOXYETHYL)-RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(C43)P*(G48)P*(C43)P*(G48)P*(A44)P*(A44)P*(U36)P*(U36)P*(C43)P*(G48)P*(C43)P*(G48))-3') | | Authors: | Teplova, M, Minasov, G, Tereshko, V, Inamati, G, Cook, P.D, Egli, M. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure and improved antisense properties of 2'-O-(2-methoxyethyl)-RNA.
Nat.Struct.Biol., 6, 1999
|
|
2G92
 
 | |
8K5S
 
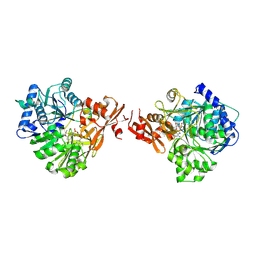 | | The structure of EntE with 3-(prop-2-yn-1-yloxy)benzoic acid sulfamoyl adenosine | | Descriptor: | Enterobactin synthase component E, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl ~{N}-(3-prop-2-ynoxyphenyl)carbonylsulfamate | | Authors: | Miyanaga, A, Ishikawa, F. | | Deposit date: | 2023-07-24 | | Release date: | 2024-07-31 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Biosynthetic Incorporation of Non-native Aryl Acid Building Blocks into Peptide Products Using Engineered Adenylation Domains.
Acs Chem.Biol., 19, 2024
|
|
8K5T
 
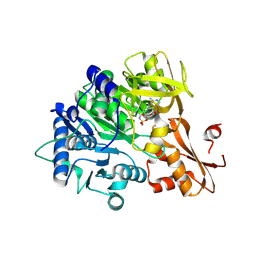 | | The structure of EntE with2-methyl-3-chloro-benzoic acid sulfamoyl adenosine | | Descriptor: | Enterobactin synthase component E, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl ~{N}-(3-chloranyl-2-methyl-phenyl)carbonylsulfamate | | Authors: | Miyanaga, A, Ishikawa, F. | | Deposit date: | 2023-07-24 | | Release date: | 2024-07-31 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biosynthetic Incorporation of Non-native Aryl Acid Building Blocks into Peptide Products Using Engineered Adenylation Domains.
Acs Chem.Biol., 19, 2024
|
|
2DDX
 
 | |
2FIL
 
 | |
2FII
 
 | |
2FIH
 
 | |
2FIJ
 
 | |
3VPL
 
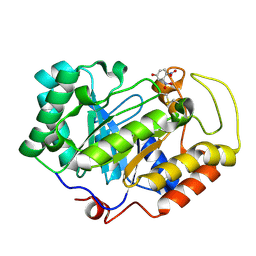 | | Crystal structure of a 2-fluoroxylotriosyl complex of the Vibrio sp. AX-4 Beta-1,3-xylanase | | Descriptor: | 3,4-dinitrophenol, Beta-1,3-xylanase XYL4, beta-D-xylopyranose-(1-3)-beta-D-xylopyranose-(1-3)-2-deoxy-2-fluoro-beta-D-xylopyranose, ... | | Authors: | Watanabe, N, Sakaguchi, K. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The crystal structure of a 2-fluoroxylotriosyl complex of the Vibrio sp. AX-4 beta-1,3-xylanase at 1.2 A resolution
To be Published
|
|
