5GHJ
 
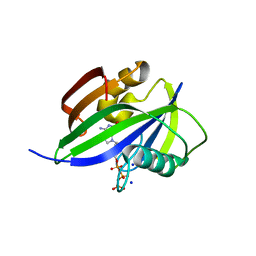 | | Crystal structure of human MTH1(G2K mutant) in complex with 2-oxo-dATP | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Waz, S, Hirata, K, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2016-06-20 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity
J. Biol. Chem., 292, 2017
|
|
5GHP
 
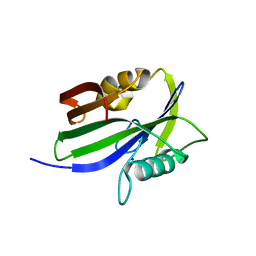 | | Crystal structure of human MTH1(G2K/D120A mutant) in complex with 2-oxo-dATP | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION | | Authors: | Nakamura, T, Waz, S, Hirata, K, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2016-06-20 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.192 Å) | | Cite: | Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity
J. Biol. Chem., 292, 2017
|
|
6LCS
 
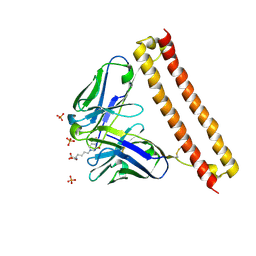 | | Crystal structure of 73MuL9 Fv-clasp fragment in complex with GA-pyridine analogue | | Descriptor: | (2~{S})-6-[4-(hydroxymethyl)-3-oxidanyl-pyridin-1-ium-1-yl]-2-(phenylmethoxycarbonylamino)hexanoic acid, PHOSPHATE ION, VH-SARAH, ... | | Authors: | Nakamura, T, Takagi, J, Yamagata, Y, Morioka, H. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular recognition of a single-chain Fv antibody specific for GA-pyridine, an advanced glycation end-product (AGE), elucidated using biophysical techniques and synthetic antigen analogues.
J.Biochem., 170, 2021
|
|
5WS7
 
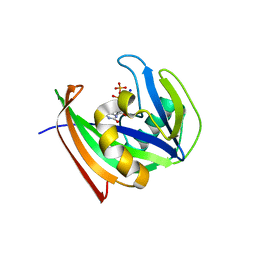 | | Crystal structure of human MTH1(G2K/C87A/C104S mutant) in complex with 2-oxo-dATP | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Waz, S, Hirata, K, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2016-12-05 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity
J. Biol. Chem., 292, 2017
|
|
5B36
 
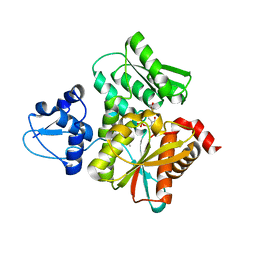 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with Cysteine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYSTEINE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Nakamura, T, Takeda, E, Kawai, Y, Kataoka, M, Ishikawa, K. | | Deposit date: | 2016-02-10 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Role of F225 in O-phosphoserine sulfhydrylase from Aeropyrum pernix K1
Extremophiles, 20, 2016
|
|
5B3A
 
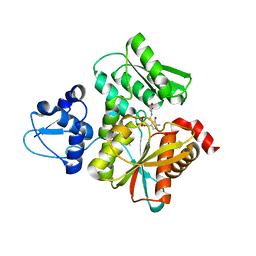 | | Crystal Structure of O-Phoshoserine Sulfhydrylase from Aeropyrum pernix in Complexed with the alpha-Aminoacrylate Intermediate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, Protein CysO | | Authors: | Nakamura, T, Takeda, E, Kawai, Y, Kataoka, M, Ishikawa, K. | | Deposit date: | 2016-02-12 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Role of F225 in O-phosphoserine sulfhydrylase from Aeropyrum pernix K1
Extremophiles, 20, 2016
|
|
2CZN
 
 | | Solution structure of the chitin-binding domain of hyperthermophilic chitinase from pyrococcus furiosus | | Descriptor: | chitinase | | Authors: | Uegaki, T, Ikegami, T, Nakamura, T, Hagihara, Y, Mine, S, Inoue, T, Matsumura, H, Ataka, M, Ishikawa, K. | | Deposit date: | 2005-07-13 | | Release date: | 2006-07-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Tertiary structure and carbohydrate recognition by the chitin-binding domain of a hyperthermophilic chitinase from Pyrococcus furiosus.
J.Mol.Biol., 381, 2008
|
|
6ILI
 
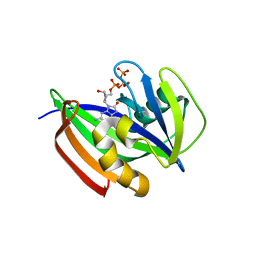 | | Crystal structure of human MTH1(G2K/D120N mutant) in complex with 8-oxo-dGTP at pH 6.5 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE | | Authors: | Nakamura, T, Waz, S, Hirata, K, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2018-10-18 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity
J. Biol. Chem., 292, 2017
|
|
3WL4
 
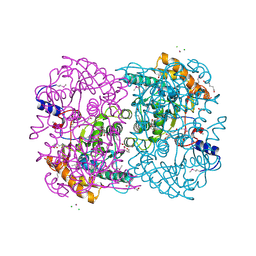 | | N,N'-diacetylchitobiose deacetylase (Se-derivative) from Pyrococcus furiosus | | Descriptor: | CADMIUM ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Nakamura, T, Niiyama, M, Hashimoto, W, Uegaki, K. | | Deposit date: | 2013-11-07 | | Release date: | 2014-05-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Expression from engineered Escherichia coli chromosome and crystallographic study of archaeal N,N'-diacetylchitobiose deacetylase
Febs J., 281, 2014
|
|
2CWR
 
 | |
3WL3
 
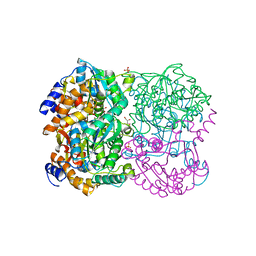 | | N,N'-diacetylchitobiose deacetylase from Pyrococcus horikoshii | | Descriptor: | GLYCEROL, PHOSPHATE ION, Putative uncharacterized protein PH0499, ... | | Authors: | Nakamura, T, Niiyama, M, Hashimoto, W, Uegaki, K. | | Deposit date: | 2013-11-07 | | Release date: | 2014-05-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Expression from engineered Escherichia coli chromosome and crystallographic study of archaeal N,N'-diacetylchitobiose deacetylase
Febs J., 281, 2014
|
|
6IU1
 
 | |
6IU0
 
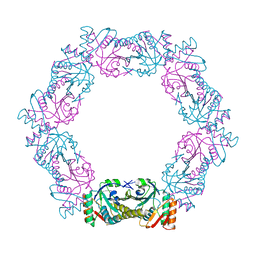 | |
6ITZ
 
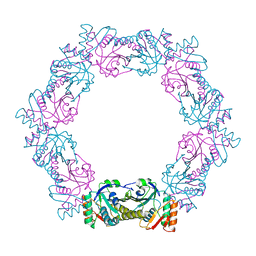 | | Peroxiredoxin from Thermococcus kodakaraensis | | Descriptor: | Peroxiredoxin | | Authors: | Nakamura, T, Himiyama, T. | | Deposit date: | 2018-11-27 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Distinct molecular assembly of homologous peroxiredoxins from Pyrococcus horikoshii and Thermococcus kodakaraensis.
J.Biochem., 166, 2019
|
|
3NG5
 
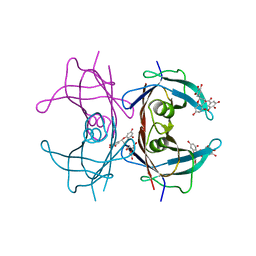 | | Crystal Structure of V30M transthyretin complexed with (-)-epigallocatechin gallate (EGCG) | | Descriptor: | (2R,3R)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromen-3-yl 3,4,5-trihydroxybenzoate, GLYCEROL, Transthyretin | | Authors: | Miyata, M, Nakamura, T, Ikemizu, S, Chirifu, M, Yamagata, Y, Kai, H. | | Deposit date: | 2010-06-11 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of green tea polyphenol(-)-epigallocatechin gallate (EGCG)-transthyretin complex reveals novel binding site distinct from thyroxine binding site
Biochemistry, 2010
|
|
7F8D
 
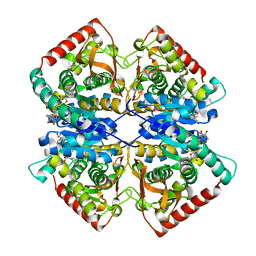 | | Malate Dehydrogenase from Geobacillus stearothermophilus (gs-MDH) G218Y mutant | | Descriptor: | Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Shimozawa, Y, Himiyama, T, Nakamura, T, Nishiya, Y. | | Deposit date: | 2021-07-02 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Increasing loop flexibility affords low-temperature adaptation of a moderate thermophilic malate dehydrogenase from Geobacillus stearothermophilus.
Protein Eng.Des.Sel., 34, 2021
|
|
5LAO
 
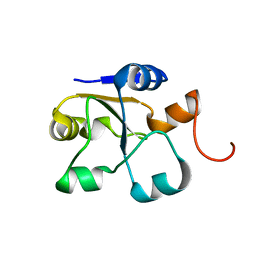 | | S-nitrosylated 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Maslennikov, I, Kwiatkowski, W, Choe, S, Lipton, S.A, Guntert, P, Riek, R. | | Deposit date: | 2016-06-14 | | Release date: | 2016-08-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
5LAM
 
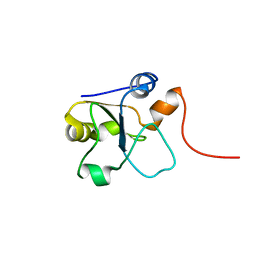 | | Refined 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Maslennikov, I, Kwiatkowski, W, Choe, S, Lipton, S.A, Guntert, P, Riek, R. | | Deposit date: | 2016-06-14 | | Release date: | 2016-08-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
6M5Y
 
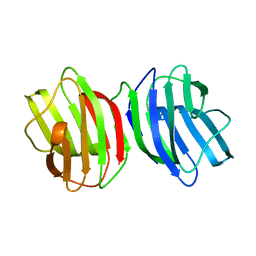 | |
5XQY
 
 | |
5XP1
 
 | |
6L0S
 
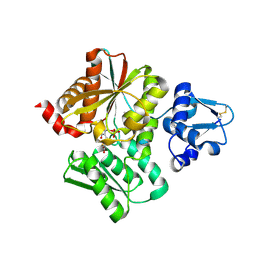 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with L-Cysteine | | Descriptor: | (2R)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-sulfanyl-propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Protein CysO | | Authors: | Nakabayashi, M, Takeda, E, Ishikawa, K, Nakamura, T. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Identification of amino acid residues important for recognition of O-phospho-l-serine substrates by cysteine synthase.
J.Biosci.Bioeng., 131, 2021
|
|
6L0Q
 
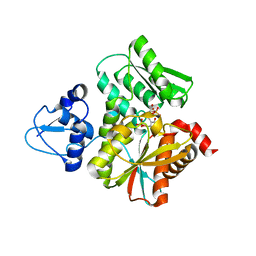 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with O-Phosphoserine | | Descriptor: | (2S)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-phosphonooxy-propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Protein CysO | | Authors: | Nakabayashi, M, Takeda, E, Ishikawa, K, Nakamura, T. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Identification of amino acid residues important for recognition of O-phospho-l-serine substrates by cysteine synthase.
J.Biosci.Bioeng., 131, 2021
|
|
6L0P
 
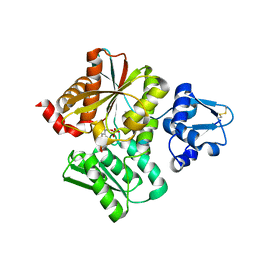 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with O-Phosphoserine | | Descriptor: | (2S)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-phosphonooxy-propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Protein CysO | | Authors: | Nakabayashi, M, Takeda, E, Ishikawa, K, Nakamura, T. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of amino acid residues important for recognition of O-phospho-l-serine substrates by cysteine synthase.
J.Biosci.Bioeng., 131, 2021
|
|
6KRS
 
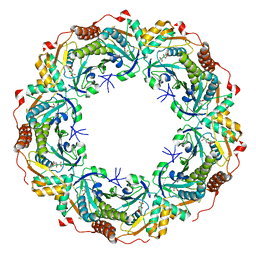 | |
