1R6T
 
 | | crystal structure of human tryptophanyl-tRNA synthetase | | Descriptor: | GLYCEROL, TRYPTOPHANYL-5'AMP, Tryptophanyl-tRNA synthetase | | Authors: | Yang, X.-L, Otero, F.J, Skene, R.J, McRee, D.E, Ribas de Pouplana, L, Schimmel, P. | | Deposit date: | 2003-10-16 | | Release date: | 2004-01-06 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures that suggest late development of genetic code components for differentiating aromatic side chains
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1Q11
 
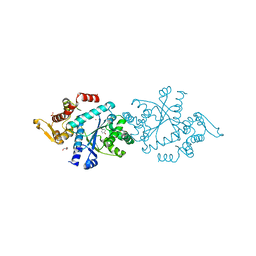 | | Crystal structure of an active fragment of human tyrosyl-tRNA synthetase with tyrosinol | | Descriptor: | 4-[(2S)-2-amino-3-hydroxypropyl]phenol, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Yang, X.-L, Schimmel, P, Ribas de Pouplana, L. | | Deposit date: | 2003-07-18 | | Release date: | 2004-01-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures that suggest late development of genetic code components for differentiating aromatic side chains
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1S62
 
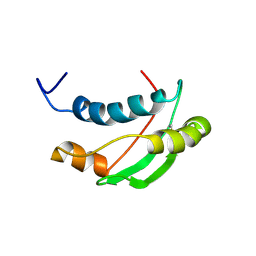 | | Solution structure of the Escherichia coli TolA C-terminal domain | | Descriptor: | TolA protein | | Authors: | Deprez, C, Blanchard, L, Simorre, J.-P, Gavioli, M, Guerlesquin, F, Lazdunski, C, Lloubes, R, Marion, D. | | Deposit date: | 2004-01-22 | | Release date: | 2005-02-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the E.coli TolA C-terminal domain reveals conformational changes upon binding to the phage g3p N-terminal domain.
J.Mol.Biol., 346, 2005
|
|
1RZT
 
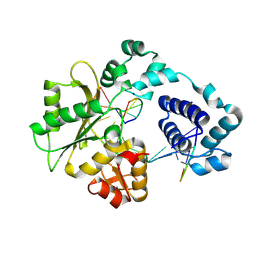 | | Crystal structure of DNA polymerase lambda complexed with a two nucleotide gap DNA molecule | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*CP*GP*GP*CP*AP*AP*CP*GP*CP*AP*C)-3', 5'-D(*GP*TP*GP*CP*G)-3', ... | | Authors: | Pedersen, L.C, Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Blanco, L, Kunkel, T.A. | | Deposit date: | 2003-12-29 | | Release date: | 2004-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural solution for the DNA polymerase lambda-dependent repair of DNA gaps with minimal homology.
Mol.Cell, 13, 2004
|
|
1RIQ
 
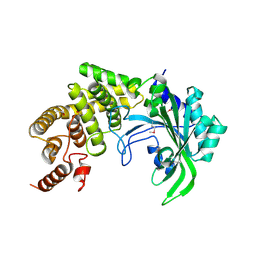 | | The crystal structure of the catalytic fragment of the alanyl-tRNA synthetase | | Descriptor: | Alanyl-tRNA synthetase | | Authors: | Swairjo, M.A, Otero, F.J, Yang, X.-L, Lovato, M.A, Skene, R.J, McRee, D.E, Ribas de Pouplana, L, Schimmel, P. | | Deposit date: | 2003-11-17 | | Release date: | 2004-04-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Alanyl-tRNA Synthetase Crystal Structure and Design for Acceptor-Stem Recognition
Mol.Cell, 13, 2004
|
|
8AIT
 
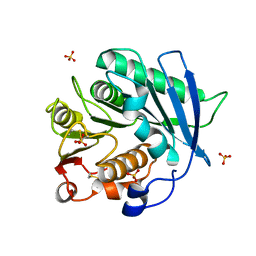 | | Crystal structure of cutinase PbauzCut from Pseudomonas bauzanensis | | Descriptor: | Cutinase, SULFATE ION | | Authors: | Zahn, M, Allen, M.D, Pickford, A.R, McGeehan, J.E. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Concentration-Dependent Inhibition of Mesophilic PETases on Poly(ethylene terephthalate) Can Be Eliminated by Enzyme Engineering.
ChemSusChem, 16, 2023
|
|
8AIS
 
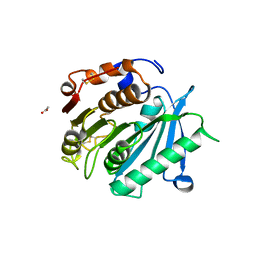 | | Crystal structure of cutinase PsCut from Pseudomonas saudimassiliensis | | Descriptor: | ACETATE ION, Lipase 1 | | Authors: | Zahn, M, Allen, M.D, Pickford, A.R, McGeehan, J.E. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Concentration-Dependent Inhibition of Mesophilic PETases on Poly(ethylene terephthalate) Can Be Eliminated by Enzyme Engineering.
ChemSusChem, 16, 2023
|
|
8AIR
 
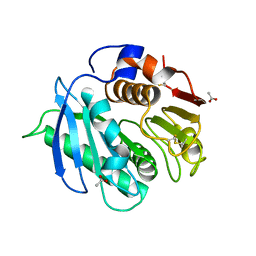 | | Crystal structure of cutinase RgCutII from Rhizobacter gummiphilus | | Descriptor: | ACETATE ION, RgCutII | | Authors: | Zahn, M, Allen, M.D, Pickford, A.R, McGeehan, J.E. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Concentration-Dependent Inhibition of Mesophilic PETases on Poly(ethylene terephthalate) Can Be Eliminated by Enzyme Engineering.
ChemSusChem, 16, 2023
|
|
7Z5G
 
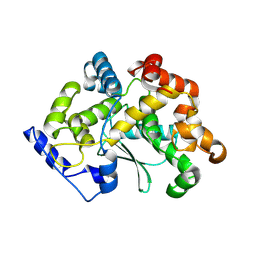 | | human apo MATCAP | | Descriptor: | Uncharacterized protein KIAA0895-like | | Authors: | Bak, J, Adamoupolos, A, Heidebrecht, T, Perrakis, A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-05-11 | | Last modified: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | Posttranslational modification of microtubules by the MATCAP detyrosinase.
Science, 376, 2022
|
|
7Z6S
 
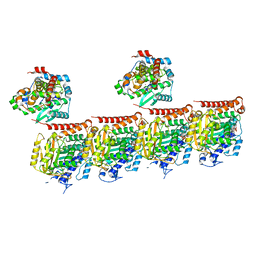 | | MATCAP bound to a human 14 protofilament microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bak, J, Perrakis, A. | | Deposit date: | 2022-03-14 | | Release date: | 2022-05-11 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Posttranslational modification of microtubules by the MATCAP detyrosinase.
Science, 376, 2022
|
|
7Z5H
 
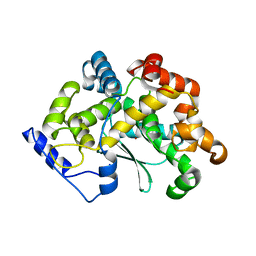 | | human Zn MATCAP | | Descriptor: | Uncharacterized protein KIAA0895-like, ZINC ION | | Authors: | Bak, J, Adamopoulos, A, Heidebrecht, T, Perrakis, A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Posttranslational modification of microtubules by the MATCAP detyrosinase.
Science, 376, 2022
|
|
8RXB
 
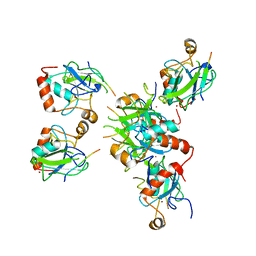 | | Human UPF1 CH domain in complex with SMG6 peptide | | Descriptor: | Regulator of nonsense transcripts 1, Telomerase-binding protein EST1A, ZINC ION | | Authors: | Langer, L, Basquin, J, Conti, E. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | UPF1 helicase orchestrates mutually exclusive interactions with the SMG6 endonuclease and UPF2.
Nucleic Acids Res., 52, 2024
|
|
5T7H
 
 | | Crystal structure of dimeric yeast iso-1-cytochrome C with CYMAL6 | | Descriptor: | 6-cyclohexylhexan-1-ol, Cytochrome c iso-1, HEME C, ... | | Authors: | Mcclelland, L, Mou, T.C, Sprang, S.R, Bowler, B.E. | | Deposit date: | 2016-09-05 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Cytochrome c Can Form a Well-Defined Binding Pocket for Hydrocarbons.
J. Am. Chem. Soc., 138, 2016
|
|
7WB5
 
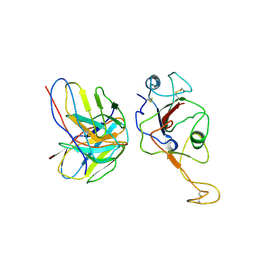 | | local structure of hu33 and spike | | Descriptor: | Surface glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, hu33 heavy chain, ... | | Authors: | Pulan, L. | | Deposit date: | 2021-12-15 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A non-ACE2-blocking neutralizing antibody against Omicron-included SARS-CoV-2 variants.
Signal Transduct Target Ther, 7, 2022
|
|
7WBH
 
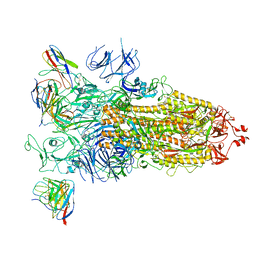 | | overall structure of hu33 and spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Pulan, L. | | Deposit date: | 2021-12-16 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A non-ACE2-blocking neutralizing antibody against Omicron-included SARS-CoV-2 variants.
Signal Transduct Target Ther, 7, 2022
|
|
7T3D
 
 | | CryoEM map of anchor 222-1C06 Fab and lateral patch 2B05 Fab binding H1 HA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 222-1C06 mAb heavy chain, ... | | Authors: | Han, J, Ward, A.B. | | Deposit date: | 2021-12-07 | | Release date: | 2022-01-12 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Broadly neutralizing antibodies target a haemagglutinin anchor epitope.
Nature, 602, 2022
|
|
4V4J
 
 | | Interactions and Dynamics of the Shine-Dalgarno Helix in the 70S Ribosome. | | Descriptor: | 16S RNA, 23S LARGE SUBUNIT RIBOSOMAL RNA, 30S ribosomal protein S10, ... | | Authors: | Korostelev, A, Trakhanov, S, Asahara, H, Laurberg, M, Noller, H.F. | | Deposit date: | 2007-07-18 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.83 Å) | | Cite: | Interactions and dynamics of the Shine Dalgarno helix in the 70S ribosome.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
5XPD
 
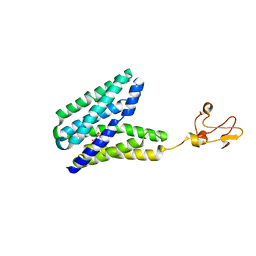 | |
5YEW
 
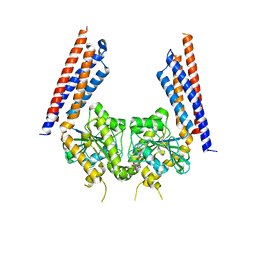 | | Structural basis for GTP hydrolysis and conformational change of mitofusin 1 in mediating mitochondrial fusion | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yan, L, Qi, Y, Huang, X, Yu, C. | | Deposit date: | 2017-09-20 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for GTP hydrolysis and conformational change of MFN1 in mediating membrane fusion
Nat. Struct. Mol. Biol., 25, 2018
|
|
6CFD
 
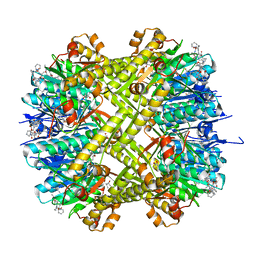 | | ADEP4 bound to E. faecium ClpP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit, N-[(6aS,12S,15aS,17R,21R,23aS)-17,21-dimethyl-6,11,15,20,23-pentaoxooctadecahydro-2H,6H,11H,15H-pyrido[2,1-i]dipyrrolo[2,1-c:2',1'-l][1,4,7,10,13]oxatetraazacyclohexadecin-12-yl]-3,5-difluoro-Nalpha-[(2E)-hept-2-enoyl]-L-phenylalaninamide | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2018-02-14 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | In VivoandIn VitroEffects of a ClpP-Activating Antibiotic against Vancomycin-Resistant Enterococci.
Antimicrob. Agents Chemother., 62, 2018
|
|
1KHQ
 
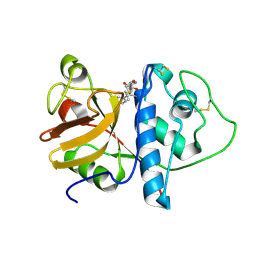 | | ORTHORHOMBIC FORM OF PAPAIN/ZLFG-DAM COVALENT COMPLEX | | Descriptor: | Papain, peptidic inhibitor | | Authors: | Janowski, R, Kozak, M, Jankowska, E, Grzonka, Z, Jaskolski, M. | | Deposit date: | 2001-11-30 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two polymorphs of a covalent complex between papain and a diazomethylketone inhibitor
J.Pept.Res., 64, 2004
|
|
1KHP
 
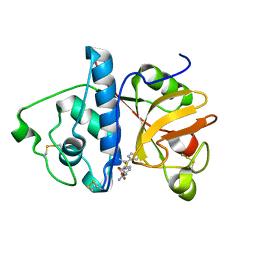 | | Monoclinic form of papain/ZLFG-DAM covalent complex | | Descriptor: | Papain, peptidic inhibitor | | Authors: | Janowski, R, Kozak, M, Jankowska, E, Grzonka, Z, Jaskolski, M. | | Deposit date: | 2001-11-30 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two polymorphs of a covalent complex between papain and a diazomethylketone inhibitor
J.Pept.Res., 64, 2004
|
|
5C90
 
 | | Staphylococcus aureus ClpP mutant - Y63A | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit | | Authors: | Ye, F, Liu, H, Zhang, J, Gan, J, Yang, C.-G. | | Deposit date: | 2015-06-26 | | Release date: | 2016-05-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization of Gain-of-Function Mutant Provides New Insights into ClpP Structure
Acs Chem.Biol., 11, 2016
|
|
5DAJ
 
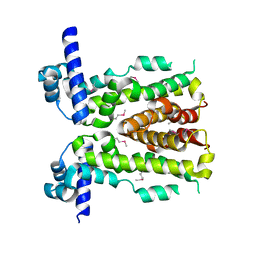 | | Crystal structure of NalD, the secondary repressor of MexAB-OprM multidrug efflux pump in Pseudomonas aeruginosa | | Descriptor: | NalD | | Authors: | Chen, W.Z, Wang, D, Huang, S.Q, Hu, Q.Y, Liu, X.C, Gan, J.H, Chen, H. | | Deposit date: | 2015-08-20 | | Release date: | 2016-04-20 | | Last modified: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Novobiocin binding to NalD induces the expression of the MexAB-OprM pump in Pseudomonas aeruginosa
Mol.Microbiol., 100, 2016
|
|
4HQE
 
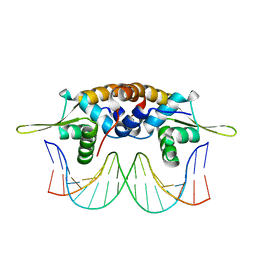 | | The crystal structure of QsrR-DNA complex | | Descriptor: | DNA (5'-D(*AP*GP*TP*AP*TP*AP*AP*TP*TP*AP*TP*TP*AP*TP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*TP*AP*AP*TP*AP*AP*TP*TP*AP*TP*AP*CP*T)-3'), Transcriptional regulator QsrR | | Authors: | He, C, Ji, Q, Zhang, L. | | Deposit date: | 2012-10-25 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Molecular mechanism of quinone signaling mediated through S-quinonization of a YodB family repressor QsrR.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
