9BCG
 
 | | Myeloid cell leukemia-1 (Mcl-1) complexed with compound | | Descriptor: | 7-[(4R,5S,6P)-7-chloro-10-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-4-methyl-1-oxo-6-(1,3,5-trimethyl-1H-pyrazol-4-yl)-3,4-dihydropyrazino[1,2-a]indol-2(1H)-yl]-4,5-dimethoxy-1-methyl-1H-indole-2-carboxylic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhao, B, Fesik, S.W. | | Deposit date: | 2024-04-09 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Discovery of a Myeloid Cell Leukemia 1 (Mcl-1) Inhibitor That Demonstrates Potent In Vivo Activities in Mouse Models of Hematological and Solid Tumors.
J.Med.Chem., 67, 2024
|
|
7UAS
 
 | |
5FC4
 
 | | Mcl-1 complexed with small molecule inhibitor | | Descriptor: | 2-[5-[1,1,2,2-tetrakis(fluoranyl)ethyl]-1~{H}-pyrazol-3-yl]phenol, 6-chloranyl-~{N}-methylsulfonyl-3-(3-naphthalen-1-yloxypropyl)-1~{H}-indole-2-carboxamide, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhao, B. | | Deposit date: | 2015-12-14 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of 2-Indole-acylsulfonamide Myeloid Cell Leukemia 1 (Mcl-1) Inhibitors Using Fragment-Based Methods.
J.Med.Chem., 59, 2016
|
|
5FDR
 
 | | Mcl-1 complexed with small molecule inhibitor | | Descriptor: | 5-[[6-chloranyl-3-[3-(4-chloranyl-3,5-dimethyl-phenoxy)propyl]-7-(3,5-dimethyl-1~{H}-pyrazol-4-yl)-1~{H}-indol-2-yl]carbonylsulfamoyl]furan-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhao, B. | | Deposit date: | 2015-12-16 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of 2-Indole-acylsulfonamide Myeloid Cell Leukemia 1 (Mcl-1) Inhibitors Using Fragment-Based Methods.
J.Med.Chem., 59, 2016
|
|
5FDO
 
 | | Mcl-1 complexed with small molecule inhibitor | | Descriptor: | 3-[3-(4-chloranyl-3,5-dimethyl-phenoxy)propyl]-~{N}-(phenylsulfonyl)-1~{H}-indole-2-carboxamide, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhao, B. | | Deposit date: | 2015-12-16 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of 2-Indole-acylsulfonamide Myeloid Cell Leukemia 1 (Mcl-1) Inhibitors Using Fragment-Based Methods.
J.Med.Chem., 59, 2016
|
|
6NOS
 
 | | PD-L1 IgV domain V76T with fragment | | Descriptor: | 1-[5-(3,5-dichlorophenyl)furan-2-yl]-N-methylmethanamine, Programmed cell death 1 ligand 1 | | Authors: | Zhao, B, Perry, E. | | Deposit date: | 2019-01-16 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Fragment-based screening of programmed death ligand 1 (PD-L1).
Bioorg. Med. Chem. Lett., 29, 2019
|
|
4HW2
 
 | | Discovery of potent Mcl-1 inhibitors using fragment-based methods and structure-based design | | Descriptor: | 1,2-ETHANEDIOL, 6-chloro-3-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1H-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | Authors: | Zhao, B. | | Deposit date: | 2012-11-07 | | Release date: | 2013-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of potent myeloid cell leukemia 1 (Mcl-1) inhibitors using fragment-based methods and structure-based design.
J.Med.Chem., 56, 2013
|
|
4HW3
 
 | | Discovery of potent Mcl-1 inhibitors using fragment-based methods and structure-based design | | Descriptor: | 3-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1-benzothiophene-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhao, B. | | Deposit date: | 2012-11-07 | | Release date: | 2013-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of potent myeloid cell leukemia 1 (Mcl-1) inhibitors using fragment-based methods and structure-based design.
J.Med.Chem., 56, 2013
|
|
4OEL
 
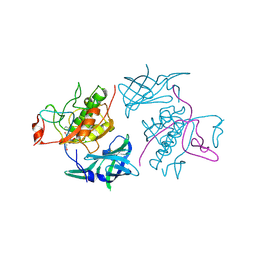 | | Crystal structure of Cathepsin C in complex with dipeptide substrates | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Dipeptidyl peptidase 1 Heavy chain, ... | | Authors: | Zhao, B, Smallwood, A, Concha, N. | | Deposit date: | 2014-01-13 | | Release date: | 2015-03-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The amino-acid substituents of dipeptide substrates of cathepsin C can determine the rate-limiting steps of catalysis.
Biochemistry, 51, 2012
|
|
4OEM
 
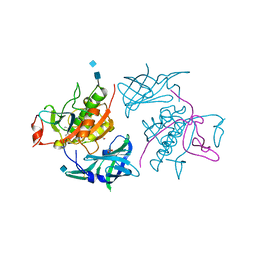 | | Crystal structure of Cathepsin C in complex with dipeptide substrates | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zhao, B, Smallwood, A, Concha, N. | | Deposit date: | 2014-01-13 | | Release date: | 2015-03-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The amino-acid substituents of dipeptide substrates of cathepsin C can determine the rate-limiting steps of catalysis.
Biochemistry, 51, 2012
|
|
1AYW
 
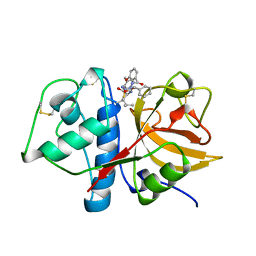 | | CRYSTAL STRUCTURE OF CYSTEINE PROTEASE HUMAN CATHEPSIN K IN COMPLEX WITH A COVALENT BENZYLOXYBENZOYLCARBOHYDRAZIDE INHIBITOR | | Descriptor: | 1-(N-BENZYLOXYCARBONYL-L-LEUCINYL)-5-(3-BENZYLOXY BENZOYL)CARBOHYDRAZIDE, CATHEPSIN K | | Authors: | Zhao, B, Smith, W.W, Janson, C.A, Abdel-Meguid, S.S. | | Deposit date: | 1997-11-10 | | Release date: | 1998-11-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design of potent and selective human cathepsin K inhibitors that span the active site.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1AU4
 
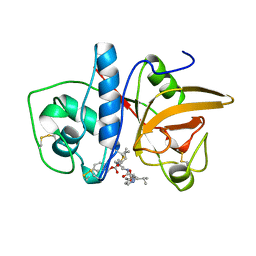 | | CRYSTAL STRUCTURE OF THE CYSTEINE PROTEASE HUMAN CATHEPSIN K IN COMPLEX WITH A COVALENT PYRROLIDINONE INHIBITOR | | Descriptor: | 4-[[N-[(PHENYLMETHOXY)CARBONYL]-/NL/N-LEUCYL]AMINO]-1[(2S)-2-[[[4-(PYRIDINYLMETHOXY)CARBONYL]AMINO]-4-METHYLPENT/NYL]-3-PYRROLIDINONE/N, CATHEPSIN K | | Authors: | Zhao, B, Smith, W.W, Janson, C.A, Abdel-Meguid, S.S. | | Deposit date: | 1997-09-10 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformationally constrained 1,3-diamino ketones: a series of potent inhibitors of the cysteine protease cathepsin K.
J.Med.Chem., 41, 1998
|
|
1AU0
 
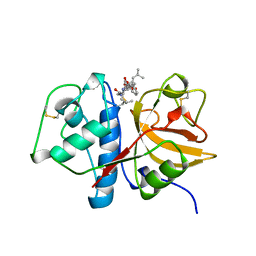 | | CRYSTAL STRUCTURE OF THE CYSTEINE PROTEASE HUMAN CATHEPSIN K IN COMPLEX WITH A COVALENT SYMMETRIC DIACYLAMINOMETHYL KETONE INHIBITOR | | Descriptor: | 1,3-BIS[[N-[(PHENYLMETHOXY)CARBONYL]-L-LEUCYL]AMINO]-2-PROPANONE, CATHEPSIN K | | Authors: | Zhao, B, Smith, W.W, Janson, C.A, Abdel-Meguid, S.S. | | Deposit date: | 1997-09-09 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Design of Potent and Selective Cathepsin K Inhibitors
J.Am.Chem.Soc., 119, 1997
|
|
1AYV
 
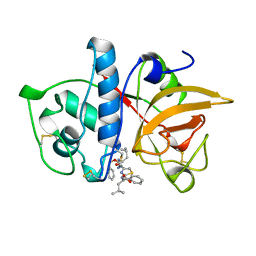 | | CRYSTAL STRUCTURE OF CYSTEINE PROTEASE HUMAN CATHEPSIN K IN COMPLEX WITH A COVALENT THIAZOLHYDRAZIDE INHIBITOR | | Descriptor: | CATHEPSIN K, N-[2-[1-(N-BENZYLOXYCARBONYLAMINO)-3-METHYLBUTYL]THIAZOL-4-YLCARBONYL]-N'-(BENZYLOXYCARBONYL-L-LEUCINYL)HYDRAZIDE | | Authors: | Zhao, B, Smith, W.W, Janson, C.A, Abdel-Meguid, S.S. | | Deposit date: | 1997-11-10 | | Release date: | 1998-11-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of potent and selective human cathepsin K inhibitors that span the active site.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1AU3
 
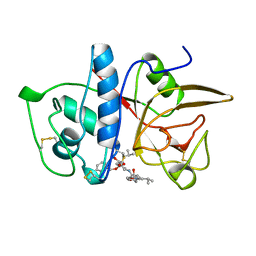 | | CRYSTAL STRUCTURE OF THE CYSTEINE PROTEASE HUMAN CATHEPSIN K IN COMPLEX WITH A COVALENT PYRROLIDINONE INHIBITOR | | Descriptor: | 1-[N[(PHENYLMETHOXY)CARBONYL]-L-LEUCYL-4-[[N/N-[(PHENYLMETHOXY)CARBONYL]-/NL-LEUCYL]AMINO]-3-PYRROLIDINONE/N, CATHEPSIN K | | Authors: | Zhao, B, Smith, W.W, Janson, C.A, Abdel-Meguid, S.S. | | Deposit date: | 1997-09-10 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformationally constrained 1,3-diamino ketones: a series of potent inhibitors of the cysteine protease cathepsin K.
J.Med.Chem., 41, 1998
|
|
1AYU
 
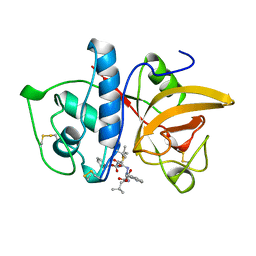 | | CRYSTAL STRUCTURE OF CYSTEINE PROTEASE HUMAN CATHEPSIN K IN COMPLEX WITH A COVALENT SYMMETRIC BISCARBOHYDRAZIDE INHIBITOR | | Descriptor: | 1,5-BIS(N-BENZYLOXYCARBONYL-L-LEUCINYL)CARBOHYDRAZIDE, CATHEPSIN K | | Authors: | Zhao, B, Smith, W.W, Janson, C.A, Abdel-Meguid, S.S. | | Deposit date: | 1997-11-10 | | Release date: | 1998-11-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design of potent and selective human cathepsin K inhibitors that span the active site.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1AU2
 
 | | CRYSTAL STRUCTURE OF THE CYSTEINE PROTEASE HUMAN CATHEPSIN K IN COMPLEX WITH A COVALENT PROPANONE INHIBITOR | | Descriptor: | 1-[[(4-PHENOXYPHENYL)SULFONYL]AMINO]-3-[[N/N-(4-PYRIDINYLCARBONYL)-L-LEUCYL]AMINO]-2-PROPANOL, CATHEPSIN K | | Authors: | Zhao, B, Smith, W.W, Janson, C.A, Abdel-Meguid, S.S. | | Deposit date: | 1997-09-10 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Design of Potent and Selective Cathepsin K Inhibitors
J.Am.Chem.Soc., 119, 1997
|
|
1DP8
 
 | |
1DP9
 
 | | CRYSTAL STRUCTURE OF IMIDAZOLE-BOUND FIXL HEME DOMAIN | | Descriptor: | FIXL PROTEIN, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gong, W, Hao, B, Chan, M.K. | | Deposit date: | 1999-12-24 | | Release date: | 2000-12-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | New mechanistic insights from structural studies of the oxygen-sensing domain of Bradyrhizobium japonicum FixL.
Biochemistry, 39, 2000
|
|
1DP6
 
 | | OXYGEN-BINDING COMPLEX OF FIXL HEME DOMAIN | | Descriptor: | FIXL PROTEIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gong, W, Hao, B, Chan, M.K. | | Deposit date: | 1999-12-23 | | Release date: | 2000-12-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New mechanistic insights from structural studies of the oxygen-sensing domain of Bradyrhizobium japonicum FixL.
Biochemistry, 39, 2000
|
|
1DFF
 
 | | PEPTIDE DEFORMYLASE | | Descriptor: | PEPTIDE DEFORMYLASE, ZINC ION | | Authors: | Chan, M.K, Gong, W, Rajagopalan, P.T.R, Hao, B, Tsai, C.M, Pei, D. | | Deposit date: | 1997-08-19 | | Release date: | 1998-09-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystal structure of the Escherichia coli peptide deformylase.
Biochemistry, 36, 1997
|
|
1DRM
 
 | | CRYSTAL STRUCTURE OF THE LIGAND FREE BJFIXL HEME DOMAIN | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SENSOR PROTEIN FIXL | | Authors: | Gong, W, Hao, B, Mansy, S.S, Gonzalez, G, Gilles-Gonzalez, M.A, Chan, M.K. | | Deposit date: | 2000-01-06 | | Release date: | 2000-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
3N54
 
 | | Crystal Structure of the GerBC protein | | Descriptor: | CHLORIDE ION, SULFATE ION, Spore germination protein B3 | | Authors: | Li, Y, Setlow, B, Setlow, P, Hao, B. | | Deposit date: | 2010-05-24 | | Release date: | 2010-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the GerBC Component of a Bacillus subtilis Spore Germinant Receptor.
J.Mol.Biol., 402, 2010
|
|
1K9X
 
 | | Structure of Pyrococcus furiosus carboxypeptidase Apo-Yb | | Descriptor: | M32 carboxypeptidase | | Authors: | Arndt, J.W, Hao, B, Ramakrishnan, V, Cheng, T, Chan, S.I, Chan, M.K. | | Deposit date: | 2001-10-31 | | Release date: | 2002-11-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of a Novel Carboxypeptidase from the Hyperthermophilic Archaeon Pyrococcus furiosus
Structure, 10, 2002
|
|
1KA2
 
 | | Structure of Pyrococcus furiosus Carboxypeptidase Apo-Mg | | Descriptor: | M32 carboxypeptidase, MAGNESIUM ION | | Authors: | Arndt, J.W, Hao, B, Ramakrishnan, V, Cheng, T, Chan, S.I, Chan, M.K. | | Deposit date: | 2001-10-31 | | Release date: | 2002-11-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Novel Carboxypeptidase from the Hyperthermophilic Archaeon Pyrococcus furiosus
Structure, 10, 2002
|
|
