1ILG
 
 | |
1K4Y
 
 | | Crystal Structure of Rabbit Liver Carboxylesterase in Complex with 4-piperidino-piperidine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-PIPERIDINO-PIPERIDINE, LIVER CARBOXYLESTERASE, ... | | Authors: | Bencharit, S, Morton, C.L, Howard-Williams, E.L, Danks, M.K, Potter, P.M, Redinbo, M.R. | | Deposit date: | 2001-10-09 | | Release date: | 2002-05-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into CPT-11 activation by mammalian carboxylesterases.
Nat.Struct.Biol., 9, 2002
|
|
1LPQ
 
 | | Human DNA Topoisomerase I (70 Kda) In Non-Covalent Complex With A 22 Base Pair DNA Duplex Containing an 8-oxoG Lesion | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*TP*(8OG)P*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3', 5'-D(*AP*AP*AP*AP*AP*TP*TP*TP*TP*TP*CP*CP*AP*AP*GP*TP*CP*TP*TP*TP*TP*T)-3', DNA topoisomerase I | | Authors: | Lesher, D.T, Pommier, Y, Stewart, L, Redinbo, M.R. | | Deposit date: | 2002-05-08 | | Release date: | 2002-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | 8-Oxoguanine rearranges the active site of human topoisomerase I
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1M13
 
 | | Crystal Structure of the Human Pregane X Receptor Ligand Binding Domain in Complex with Hyperforin, a Constituent of St. John's Wort | | Descriptor: | 4-HYDROXY-5-ISOBUTYRYL-6-METHYL-1,3,7-TRIS-(3-METHYL-BUT-2-ENYL)-6-(4-METHYL-PENT-3-ENYL)-BICYCLO[3.3.1]NON-3-ENE-2,9-DIONE, Orphan Nuclear Receptor PXR | | Authors: | Watkins, R.E, Maglich, J.M, Moore, L.B, Wisely, G.B, Noble, S.M, Davis-Searles, P.R, Lambert, M.H, Kliewer, S.A, Redinbo, M.R. | | Deposit date: | 2002-06-17 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.1 A Crystal Structure of Human PXR in Complex with the St. John's Wort Compound Hyperforin
Biochemistry, 42, 2003
|
|
4J5X
 
 | | Crystal Structure of the SR12813-bound PXR/RXRalpha LBD Heterotetramer Complex | | Descriptor: | Nuclear receptor subfamily 1 group I member 2, Nuclear receptor coactivator 1, Retinoic acid receptor RXR-alpha, ... | | Authors: | Wallace, B.D, Betts, L, Redinbo, M.R. | | Deposit date: | 2013-02-10 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Functional Analysis of the Human Nuclear Xenobiotic Receptor PXR in Complex with RXRalpha.
J.Mol.Biol., 425, 2013
|
|
6MVH
 
 | |
6MVF
 
 | |
6MVG
 
 | |
6NZG
 
 | | Bacteroides uniformis beta-glucuronidase 2 covalently bound to cyclophellitol-6-carboxylate aziridine | | Descriptor: | (1S,2R,3S,4S,5S,6R)-2-amino-3,4,5,6-tetrahydroxycyclohexane-1-carboxylic acid, Beta-galactosidase, CALCIUM ION, ... | | Authors: | Pellock, S.J, Jariwala, P.B, Redinbo, M.R. | | Deposit date: | 2019-02-13 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovering the Microbial Enzymes Driving Drug Toxicity with Activity-Based Protein Profiling.
Acs Chem.Biol., 15, 2020
|
|
6NCX
 
 | |
6NCY
 
 | | Crystal structure of hybrid beta-glucuronidase/beta-galacturonidase from Fusicatenibacter saccharivorans | | Descriptor: | Beta-glucuronidase, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Walton, W.G, Pellock, S.J, Redinbo, M.R. | | Deposit date: | 2018-12-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Selecting a Single Stereocenter: The Molecular Nuances That Differentiate beta-Hexuronidases in the Human Gut Microbiome.
Biochemistry, 58, 2019
|
|
4JKM
 
 | |
6NCZ
 
 | |
6NCW
 
 | |
4JKK
 
 | |
4JHZ
 
 | | Structure of E. coli beta-Glucuronidase bound with a novel, potent inhibitor 2-[4-(1,3-benzodioxol-5-ylmethyl)piperazin-1-yl]-N-[(1S,2S,5S)-2,5-dimethoxycyclohexyl]acetamide | | Descriptor: | 2-[4-(1,3-benzodioxol-5-ylmethyl)piperazin-1-yl]-N-[(1S,2S,5S)-2,5-dimethoxycyclohexyl]acetamide, Beta-glucuronidase | | Authors: | Roberts, A.B, Wallace, B.D, Redinbo, M.R. | | Deposit date: | 2013-03-05 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.831 Å) | | Cite: | Molecular Insights into Microbial beta-Glucuronidase Inhibition to Abrogate CPT-11 Toxicity.
Mol.Pharmacol., 84, 2013
|
|
4JKL
 
 | |
4J5W
 
 | | Crystal Structure of the apo-PXR/RXRalpha LBD Heterotetramer Complex | | Descriptor: | MAGNESIUM ION, Nuclear receptor subfamily 1 group I member 2, Nuclear receptor coactivator 1, ... | | Authors: | Wallace, B.D, Betts, L, Redinbo, M.R. | | Deposit date: | 2013-02-10 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Functional Analysis of the Human Nuclear Xenobiotic Receptor PXR in Complex with RXRalpha.
J.Mol.Biol., 425, 2013
|
|
2O6L
 
 | |
2O9I
 
 | | Crystal Structure of the Human Pregnane X Receptor LBD in complex with an SRC-1 coactivator peptide and T0901317 | | Descriptor: | N-(2,2,2-TRIFLUOROETHYL)-N-{4-[2,2,2-TRIFLUORO-1-HYDROXY-1-(TRIFLUOROMETHYL)ETHYL]PHENYL}BENZENESULFONAMIDE, Nuclear Receptor Coactivator 1 isoform 3, Orphan nuclear receptor PXR | | Authors: | Xue, Y, Redinbo, M.R. | | Deposit date: | 2006-12-13 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the PXR-T1317 complex provides a scaffold to examine the potential for receptor antagonism.
Bioorg.Med.Chem., 15, 2007
|
|
2Q7U
 
 | |
2Q1H
 
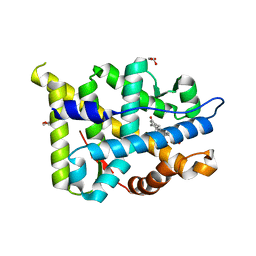 | | Ancestral Corticoid Receptor in Complex with Aldosterone | | Descriptor: | ALDOSTERONE, AncCR, GLYCEROL, ... | | Authors: | Ortlund, E.A, Bridgham, J.T, Redinbo, M.R, Thornton, J.W. | | Deposit date: | 2007-05-24 | | Release date: | 2007-09-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an ancient protein: evolution by conformational epistasis.
Science, 317, 2007
|
|
2Q1V
 
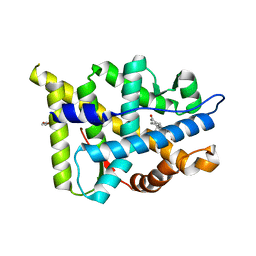 | | Ancestral corticoid receptor in complex with cortisol | | Descriptor: | 17,21-DIHYDROXYPREGNA-1,4-DIENE-3,11,20-TRIONE, AncCR, GLYCEROL | | Authors: | Ortlund, E.A, Redinbo, M.R. | | Deposit date: | 2007-05-25 | | Release date: | 2007-09-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of an ancient protein: evolution by conformational epistasis.
Science, 317, 2007
|
|
2Q3Y
 
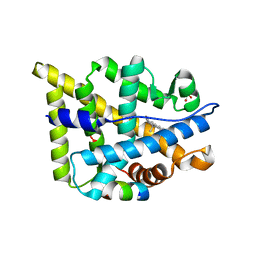 | | Ancestral Corticiod Receptor in Complex with DOC | | Descriptor: | Ancestral Corticiod Receptor, DESOXYCORTICOSTERONE, GLYCEROL, ... | | Authors: | Ortlund, E.A, Bridgham, J.T, Redinbo, M.R, Thornton, J.W. | | Deposit date: | 2007-05-30 | | Release date: | 2007-09-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an ancient protein: evolution by conformational epistasis
Science, 317, 2007
|
|
2Q7T
 
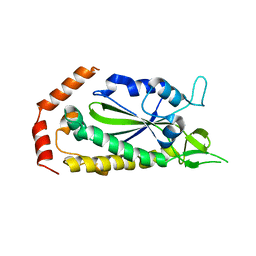 | |
