1BAV
 
 | |
7E65
 
 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3 | | Descriptor: | (2S)-2-acetamido-N-[(2R)-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]-3-(4-sulfamoylphenyl)propanamide, Peptidase M23, ZINC ION | | Authors: | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E60
 
 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 1 | | Descriptor: | (2~{R},6~{S})-2,6-diacetamido-7-[[(2~{R})-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-7-oxidanylidene-heptanoic acid, Peptidase M23, ZINC ION | | Authors: | Min, K, Yoon, H.J, Choi, Y, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E69
 
 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3-3 | | Descriptor: | N-oxidanyl-4-[(4-sulfamoylphenyl)methyl]benzamide, Peptidase M23, ZINC ION | | Authors: | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E61
 
 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 2 | | Descriptor: | 2-[[(3S)-3-acetamido-4-[[(2R)-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-4-oxidanylidene-butyl]-(phenylmethyl)amino]ethanoic acid, Peptidase M23, ZINC ION | | Authors: | Min, K.J, Yoon, H.J, Choi, Y, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E64
 
 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 2-2 | | Descriptor: | 2-[[(3S)-3-acetamido-4-[[(2R)-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-4-oxidanylidene-butyl]amino]ethanoic acid, Peptidase M23, ZINC ION | | Authors: | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E67
 
 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3-2 | | Descriptor: | N-oxidanyl-2-[4-(4-sulfamoylphenyl)phenyl]ethanamide, Peptidase M23, ZINC ION | | Authors: | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E66
 
 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3-1 | | Descriptor: | N-[2-(oxidanylamino)-2-oxidanylidene-ethyl]-2-(4-sulfamoylphenyl)ethanamide, Peptidase M23, ZINC ION | | Authors: | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E63
 
 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 2-1 | | Descriptor: | 2-[[(3S)-3-acetamido-4-[[(2R)-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-4-oxidanylidene-butyl]-(cyclopentylmethyl)amino]ethanoic acid, Peptidase M23, ZINC ION | | Authors: | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
6FCU
 
 | | The X-ray Structure of Lytic Transglycosylase Slt inactive mutant E503Q from Pseudomonas aeruginosa in complex with 4(NAG-NAMpentapeptide) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-3-O-[(2R)-1-amino-1-oxopropan-2-yl]-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, ACETATE ION, ALANINE, ... | | Authors: | Batuecas, M.T, Dominguez-Gil, T, Hermoso, J.A. | | Deposit date: | 2017-12-21 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Exolytic and endolytic turnover of peptidoglycan by lytic transglycosylase Slt ofPseudomonas aeruginosa.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FC4
 
 | |
6FCQ
 
 | |
6FCS
 
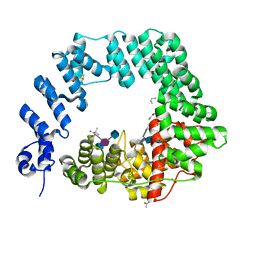 | | The X-ray Structure of Lytic Transglycosylase Slt inactive mutant E503Q from Pseudomonas aeruginosa in complex with NAG-NAMpentapeptide-NAG-NAMpentapeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-3-O-[(2R)-1-amino-1-oxopropan-2-yl]-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, ACETATE ION, ... | | Authors: | Batuecas, M.T, Dominguez-Gil, T, Hermoso, J.A. | | Deposit date: | 2017-12-21 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Exolytic and endolytic turnover of peptidoglycan by lytic transglycosylase Slt ofPseudomonas aeruginosa.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FCR
 
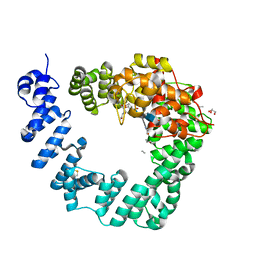 | | The X-ray Structure of Lytic Transglycosylase Slt inactive mutant E503Q from Pseudomonas aeruginosa in complex with NAG-NAMtetrapeptide-NAG-anhNAMtetrapeptide | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Batuecas, M.T, Dominguez-Gil, T, Hermoso, J.A. | | Deposit date: | 2017-12-21 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Exolytic and endolytic turnover of peptidoglycan by lytic transglycosylase Slt ofPseudomonas aeruginosa.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FBT
 
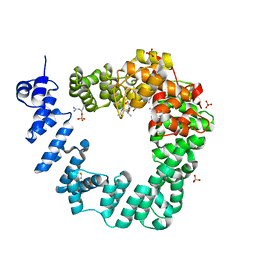 | | The X-ray Structure of Lytic Transglycosylase Slt from Pseudomonas aeruginosa in complex with the reaction product NAG-anhNAMpentapeptide | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, NAG-anhNAMpentapeptide, ... | | Authors: | Batuecas, M.T, Dominguez-Gil, T, Hermoso, J.A. | | Deposit date: | 2017-12-19 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Exolytic and endolytic turnover of peptidoglycan by lytic transglycosylase Slt ofPseudomonas aeruginosa.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4Y4V
 
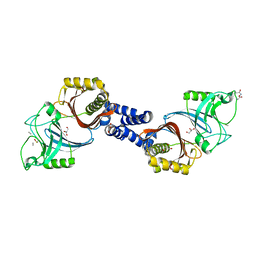 | | Structure of Helicobacter pylori Csd6 in the D-Ala-bound state | | Descriptor: | Conserved hypothetical secreted protein, D-ALANINE, GLYCEROL | | Authors: | Kim, H.S, Im, H.N, Yoon, H.J, Suh, S.W. | | Deposit date: | 2015-02-11 | | Release date: | 2015-09-02 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The Cell Shape-determining Csd6 Protein from Helicobacter pylori Constitutes a New Family of l,d-Carboxypeptidase
J.Biol.Chem., 290, 2015
|
|
2EAV
 
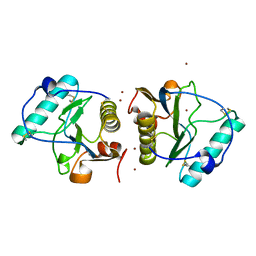 | |
2EAX
 
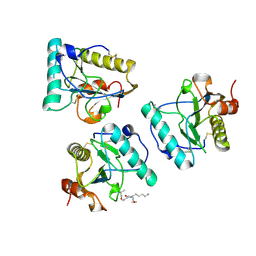 | | Crystal structure of human PGRP-IBETAC in complex with glycosamyl muramyl pentapeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside, GLYCOSAMYL MURAMYL PENTAPEPTIDE, Peptidoglycan recognition protein-I-beta | | Authors: | Cho, S. | | Deposit date: | 2007-02-03 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the bactericidal mechanism of human peptidoglycan recognition proteins
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2IXV
 
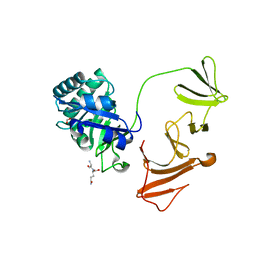 | |
2J8G
 
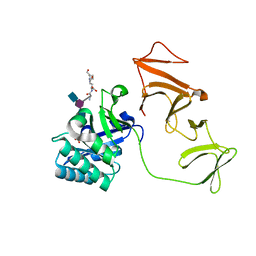 | |
2J8F
 
 | |
2IXU
 
 | |
2WWD
 
 | | 3D-structure of the modular autolysin LytC from Streptococcus pneumoniae in complex with pneummococcal peptidoglycan fragment | | Descriptor: | 1,4-BETA-N-ACETYLMURAMIDASE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-alpha-muramic acid, ALANINE, ... | | Authors: | Perez-Dorado, I, Sanles, R, Hermoso, J.A, Gonzalez, A, Garcia, A, Garcia, P, Garcia, J.L. | | Deposit date: | 2009-10-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Insights Into Pneumococcal Fratricide from the Crystal Structures of the Modular Killing Factor Lytc.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2WW5
 
 | | 3D-structure of the modular autolysin LytC from Streptococcus pneumoniae at 1.6 A resolution | | Descriptor: | 1,4-BETA-N-ACETYLMURAMIDASE, CHLORIDE ION, CHOLINE ION, ... | | Authors: | Perez-Dorado, I, Sanles, R, Hermoso, J.A, Gonzalez, A, Garcia, A, Garcia, P, Garcia, J.L, Menendez, M. | | Deposit date: | 2009-10-21 | | Release date: | 2010-04-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Insights Into Pneumococcal Fratricide from the Crystal Structures of the Modular Killing Factor Lytc.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2WWC
 
 | | 3D-structure of the modular autolysin LytC from Streptococcus pneumoniae in complex with synthetic peptidoglycan ligand | | Descriptor: | 1,4-BETA-N-ACETYLMURAMIDASE, CHOLINE ION, GLYCEROL | | Authors: | Perez-Dorado, I, Sanles, R, Hermoso, J.A, Gonzalez, A, Garcia, A, Garcia, P, Garcia, J.L. | | Deposit date: | 2009-10-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights Into Pneumococcal Fratricide from the Crystal Structures of the Modular Killing Factor Lytc.
Nat.Struct.Mol.Biol., 17, 2010
|
|
