7XF5
 
 | | Full length human CLC-2 channel in apo state | | Descriptor: | Chloride channel protein 2 | | Authors: | Wang, L. | | Deposit date: | 2022-04-01 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of ClC-2 chloride channel reveal the blocking mechanism of its specific inhibitor AK-42
Nat Commun, 14, 2023
|
|
7XMK
 
 | | Crystal structure of human RIPK1 kinase domain in complex with compound SKLB923 | | Descriptor: | 5-[2-(cyclopropylcarbonylamino)-[1,2,4]triazolo[1,5-a]pyridin-7-yl]-N-[(1S)-1-(3-fluorophenyl)ethyl]-1-methyl-indole-3-carboxamide, IODIDE ION, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Zhang, L, Wang, Y, Li, Y, Yang, S. | | Deposit date: | 2022-04-26 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.376 Å) | | Cite: | From Hit to Lead: Structure-Based Optimization of Novel Selective Inhibitors of Receptor-Interacting Protein Kinase 1 (RIPK1) for the Treatment of Inflammatory Diseases.
J.Med.Chem., 67, 2024
|
|
7XJA
 
 | | TMD masked refine map of human ClC-2 | | Descriptor: | Chloride channel protein 2 | | Authors: | Wang, L. | | Deposit date: | 2022-04-15 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of ClC-2 chloride channel reveal the blocking mechanism of its specific inhibitor AK-42
Nat Commun, 14, 2023
|
|
3DO4
 
 | |
1FVQ
 
 | | SOLUTION STRUCTURE OF THE YEAST COPPER TRANSPORTER DOMAIN CCC2A IN THE APO AND CU(I) LOADED STATES | | Descriptor: | COPPER-TRANSPORTING ATPASE | | Authors: | Banci, L, Bertini, I, Ciofi Baffoni, S, Huffman, D.L, O'Halloran, T.V. | | Deposit date: | 2000-09-20 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the yeast copper transporter domain Ccc2a in the apo and Cu(I)-loaded states.
J.Biol.Chem., 276, 2001
|
|
1FZJ
 
 | | MHC CLASS I NATURAL MUTANT H-2KBM1 HEAVY CHAIN COMPLEXED WITH BETA-2 MICROGLOBULIN AND VESICULAR STOMATITIS VIRUS NUCLEOPROTEIN | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rudolph, M.G, Speir, J.A, Brunmark, A, Mattsson, N, Jackson, M.R, Peterson, P.A, Teyton, L, Wilson, I.A. | | Deposit date: | 2000-10-03 | | Release date: | 2001-03-28 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of K(bm1) and K(bm8) reveal that subtle changes in the peptide environment impact thermostability and alloreactivity.
Immunity, 14, 2001
|
|
3M6D
 
 | | The crystal structure of the d307a mutant of glycoside Hydrolase (family 31) from ruminococcus obeum atcc 29174 | | Descriptor: | Uncharacterized protein | | Authors: | Tan, K, Tesar, C, Freeman, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-15 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Novel alpha-glucosidase from human gut microbiome: substrate specificities and their switch.
Faseb J., 24, 2010
|
|
7YE9
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
1EVF
 
 | | CRYSTAL STRUCTURE ANALYSIS OF CYS167 MUTANT OF ESCHERICHIA COLI | | Descriptor: | SULFATE ION, THYMIDYLATE SYNTHASE | | Authors: | Phan, J, Mahdavian, E, Nivens, M.C, Minor, W, Berger, S, Spencer, H.T, Dunlap, R.B, Lebioda, L. | | Deposit date: | 2000-04-19 | | Release date: | 2000-05-03 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic cysteine of thymidylate synthase is activated upon substrate binding.
Biochemistry, 39, 2000
|
|
1EWA
 
 | | Dehaloperoxidase and 4-iodophenol | | Descriptor: | 4-IODOPHENOL, DEHALOPEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | LaCount, M.W, Zhang, E, Chen, Y.P, Han, K, Whitton, M.M, Lincoln, D.E, Woodin, S.A, Lebioda, L. | | Deposit date: | 2000-04-24 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure and amino acid sequence of dehaloperoxidase from Amphitrite ornata indicate common ancestry with globins
J.Biol.Chem., 275, 2000
|
|
1EEQ
 
 | | M4L/Y(27D)D/T94H Mutant of LEN | | Descriptor: | KAPPA-4 IMMUNOGLOBULIN (LIGHT CHAIN) | | Authors: | Pokkuluri, P.R, Raffen, R, Dieckman, L, Boogaard, C, Stevens, F.J, Schiffer, M. | | Deposit date: | 2000-02-01 | | Release date: | 2001-02-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Increasing protein stability by polar surface residues: domain-wide consequences of interactions within a loop.
Biophys.J., 82, 2002
|
|
1EF7
 
 | | CRYSTAL STRUCTURE OF HUMAN CATHEPSIN X | | Descriptor: | CATHEPSIN X | | Authors: | Guncar, G, Klemencic, I, Turk, B, Turk, V, Karaoglanovic-Carmona, A, Juliano, L, Turk, D. | | Deposit date: | 2000-02-07 | | Release date: | 2000-03-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of cathepsin X: a flip-flop of the ring of His23 allows carboxy-monopeptidase and carboxy-dipeptidase activity of the protease.
Structure Fold.Des., 8, 2000
|
|
7D42
 
 | |
7D6H
 
 | |
1EVV
 
 | |
1EWZ
 
 | | CRYSTAL STRUCTURE OF THE OXA-10 BETA-LACTAMASE FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | BETA LACTAMASE OXA-10 | | Authors: | Golemi, D, Maveyraud, L, Vakulenko, S, Tranier, S, Ishiwata, A, Kotra, L.P, Samama, J.P, Mobashery, S. | | Deposit date: | 2000-04-28 | | Release date: | 2000-11-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The First Structural and Mechanistic Insights for Class D beta-Lactamases: Evidence for a Novel Catalytic Process for Turnover of beta-Lactam Antibiotics
J.Am.Chem.Soc., 122, 2000
|
|
1GBR
 
 | | ORIENTATION OF PEPTIDE FRAGMENTS FROM SOS PROTEINS BOUND TO THE N-TERMINAL SH3 DOMAIN OF GRB2 DETERMINED BY NMR SPECTROSCOPY | | Descriptor: | GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2, SOS-A PEPTIDE | | Authors: | Wittekind, M, Mapelli, C, Farmer, B.T, Suen, K.-L, Goldfarb, V, Tsao, J, Lavoie, T, Barbacid, M, Meyers, C.A, Mueller, L. | | Deposit date: | 1994-08-12 | | Release date: | 1995-01-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Orientation of peptide fragments from Sos proteins bound to the N-terminal SH3 domain of Grb2 determined by NMR spectroscopy.
Biochemistry, 33, 1994
|
|
1GJS
 
 | | Solution structure of the Albumin binding domain of Streptococcal Protein G | | Descriptor: | IMMUNOGLOBULIN G BINDING PROTEIN G | | Authors: | Johansson, M.U, Frick, I.M, Nilsson, H, Kraulis, P.J, Hober, S, Jonasson, P, Nygren, A.P, Uhlen, M, Bjorck, L, Drakenberg, T, Forsen, S, Wikstrom, M. | | Deposit date: | 2001-08-02 | | Release date: | 2001-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, Specificity, and Mode of Interaction for Bacterial Albumin-Binding Modules
J.Biol.Chem., 277, 2002
|
|
1FZO
 
 | | MHC CLASS I NATURAL MUTANT H-2KBM8 HEAVY CHAIN COMPLEXED WITH BETA-2 MICROGLOBULIN AND SENDAI VIRUS NUCLEOPROTEIN | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rudolph, M.G, Speir, J.A, Brunmark, A, Mattsson, N, Jackson, M.R, Peterson, P.A, Teyton, L, Wilson, I.A. | | Deposit date: | 2000-10-03 | | Release date: | 2001-03-28 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structures of K(bm1) and K(bm8) reveal that subtle changes in the peptide environment impact thermostability and alloreactivity.
Immunity, 14, 2001
|
|
1G00
 
 | |
1GMY
 
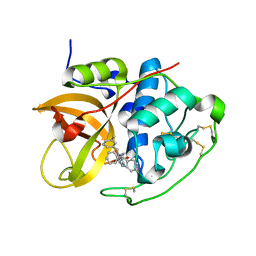 | | Cathepsin B complexed with dipeptidyl nitrile inhibitor | | Descriptor: | 2-AMINOETHANIMIDIC ACID, 3-METHYLPHENYLALANINE, CATHEPSIN B, ... | | Authors: | Greenspan, P.D, Clark, K.L, Tommasi, R.A, Cowen, S.D, McQuire, L.W, Farley, D.L, van Duzer, J.H, Goldberg, R.L, Zhou, H, Du, Z, Fitt, J.J, Coppa, D.E, Fang, Z, Macchia, W, Zhu, L, Capparelli, M.P, Goldstein, R, Wigg, A.M, Doughty, J.R, Bohacek, R.S, Knap, A.K. | | Deposit date: | 2001-09-25 | | Release date: | 2002-09-19 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of Dipeptidyl Nitriles as Potent and Selective Inhibitors of Cathepsin B Through Structure-Based Drug Design
J.Med.Chem., 44, 2001
|
|
1G98
 
 | |
7XSR
 
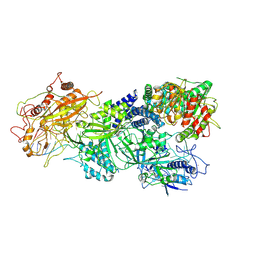 | | Structure of Craspase-target RNA | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (34-MER), ... | | Authors: | Feng, Y, Zhang, L. | | Deposit date: | 2022-05-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Target RNA activates the protease activity of Craspase to confer antiviral defense.
Mol.Cell, 82, 2022
|
|
7XSQ
 
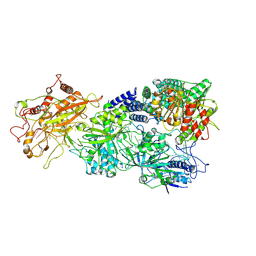 | | Structure of the Craspase | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (34-MER), ... | | Authors: | Feng, Y, Zhang, L. | | Deposit date: | 2022-05-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Target RNA activates the protease activity of Craspase to confer antiviral defense.
Mol.Cell, 82, 2022
|
|
7XT4
 
 | | Structure of Craspase-NTR | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (34-MER), ... | | Authors: | Feng, Y, Zhang, L. | | Deposit date: | 2022-05-16 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Target RNA activates the protease activity of Craspase to confer antiviral defense.
Mol.Cell, 82, 2022
|
|
