5TGN
 
 | | Crystal structure of protein Sthe_2403 from Sphaerobacter thermophilus | | Descriptor: | CHLORIDE ION, GLYCEROL, Uncharacterized protein | | Authors: | Michalska, K, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-28 | | Release date: | 2016-10-26 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure of protein Sthe_2403 from Sphaerobacter thermophilus
To Be Published
|
|
8K5R
 
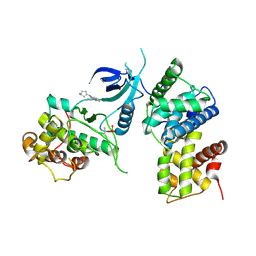 | | CDK9/cyclin T1 in complex with KB-0742 | | Descriptor: | (1S,3S)-N3-(5-pentan-3-ylpyrazolo[1,5-a]pyrimidin-7-yl)cyclopentane-1,3-diamine, Cyclin-T1, Cyclin-dependent kinase 9 | | Authors: | Zhou, M, Li, H, Gao, H, Trotter, B.W, Freeman, D. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.751 Å) | | Cite: | Discovery of KB-0742, a Potent, Selective, Orally Bioavailable Small Molecule Inhibitor of CDK9 for MYC-Dependent Cancers.
J.Med.Chem., 66, 2023
|
|
5UFH
 
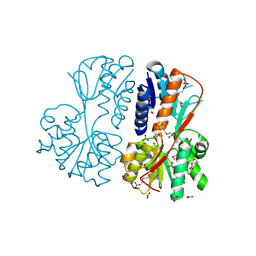 | | The crystal structure of a LacI-type transcription regulator from Bifidobacterium animalis subsp. lactis DSM 10140 | | Descriptor: | GLYCEROL, LacI-type transcriptional regulator, NITRATE ION | | Authors: | Tan, K, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-04 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The crystal structure of a LacI-type transcription regulator from Bifidobacterium animalis subsp. lactis DSM 10140
To Be Published
|
|
5UHJ
 
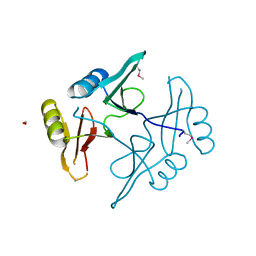 | | The crystal structure of a natural product biosynthetic enzyme from Streptomyces sp. CB03234 | | Descriptor: | FORMIC ACID, Glyoxalase/bleomycin resisance protein/dioxygenase | | Authors: | Tan, K, Li, H, Endres, M, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-01-11 | | Release date: | 2017-01-25 | | Last modified: | 2020-09-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of a natural product biosynthetic enzyme from Streptomyces sp. CB03234
To Be Published
|
|
4OVJ
 
 | |
5UJM
 
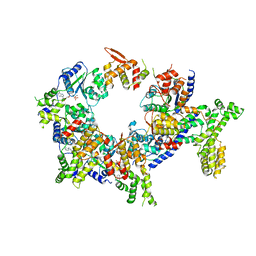 | | Structure of the active form of human Origin Recognition Complex and its ATPase motor module | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Origin recognition complex subunit 1, ... | | Authors: | Tocilj, A, On, K, Yuan, Z, Sun, J, Elkayam, E, Li, H, Stillman, B, Joshua-Tor, L. | | Deposit date: | 2017-01-18 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Structure of the active form of human Origin Recognition Complex and its ATPase motor module.
Elife, 6, 2017
|
|
5UJP
 
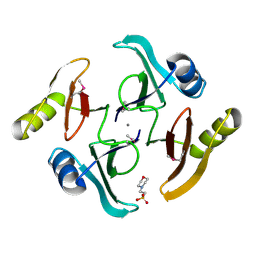 | | The crystal structure of a glyoxalase/bleomycin resistance protein from Streptomyces sp. CB03234 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Glyoxalase/bleomycin resisance protein/dioxygenase | | Authors: | Tan, K, Li, H, Endres, M, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-01-18 | | Release date: | 2017-02-22 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The crystal structure of a glyoxalase/bleomycin resistance protein from Streptomyces sp. CB03234
To Be Published
|
|
5UOB
 
 | | Structure of human endothelial nitric oxide synthase heme domain in complex with (R)-3-[(2-amino-4-methylquinolin-7-yl)methoxy]-5-(2-(methylamino)propyl)benzonitrile | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[(2-amino-4-methylquinolin-7-yl)methoxy]-5-[(2R)-2-(methylamino)propyl]benzonitrile, CHLORIDE ION, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2017-01-31 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Nitrile in the Hole: Discovery of a Small Auxiliary Pocket in Neuronal Nitric Oxide Synthase Leading to the Development of Potent and Selective 2-Aminoquinoline Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UOU
 
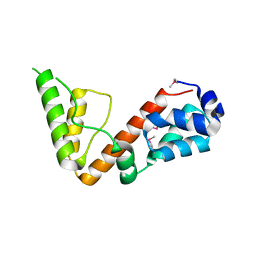 | | High resolution structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase from Klebsiella pneumoniae subsp. pneumoniae MGH 78578 | | Descriptor: | 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline (OHCU) decarboxylase | | Authors: | Chang, C, Li, H, Bearden, J, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-01 | | Release date: | 2017-02-15 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High resolution structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase from Klebsiella pneumoniae subsp. pneumoniae MGH 78578
To Be Published
|
|
5URA
 
 | | Enantiomer-Specific Binding of the Potent Antinociceptive Agent SBFI-26 to Anandamide transporters FABP7 | | Descriptor: | (1S,2S,3S,4S)-3-{[(naphthalen-1-yl)oxy]carbonyl}-2,4-diphenylcyclobutane-1-carboxylic acid, Fatty acid-binding protein, brain, ... | | Authors: | Hsu, H.-C, Li, H. | | Deposit date: | 2017-02-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85002172 Å) | | Cite: | The Antinociceptive Agent SBFI-26 Binds to Anandamide Transporters FABP5 and FABP7 at Two Different Sites.
Biochemistry, 56, 2017
|
|
5UOA
 
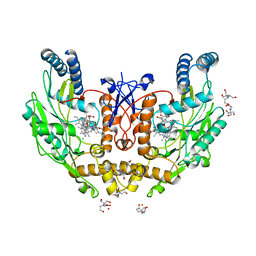 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 3-[(2-Amino-4-methylquinolin-7-yl)methoxy]-5-((methylamino)methyl)benzonitrile | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[(2-amino-4-methylquinolin-7-yl)methoxy]-5-[(methylamino)methyl]benzonitrile, CHLORIDE ION, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2017-01-31 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nitrile in the Hole: Discovery of a Small Auxiliary Pocket in Neuronal Nitric Oxide Synthase Leading to the Development of Potent and Selective 2-Aminoquinoline Inhibitors.
J. Med. Chem., 60, 2017
|
|
5V8F
 
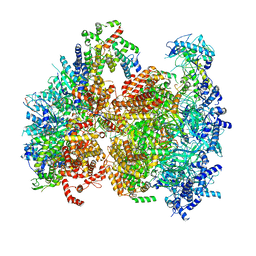 | | Structural basis of MCM2-7 replicative helicase loading by ORC-Cdc6 and Cdt1 | | Descriptor: | Cell division control protein 6, Cell division cycle protein CDT1, DNA (39-MER), ... | | Authors: | Yuan, Z, Riera, A, Bai, L, Sun, J, Spanos, C, Chen, Z.A, Barbon, M, Rappsilber, J, Stillman, B, Speck, C, Li, H. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of Mcm2-7 replicative helicase loading by ORC-Cdc6 and Cdt1.
Nat. Struct. Mol. Biol., 24, 2017
|
|
1HV8
 
 | |
6MWY
 
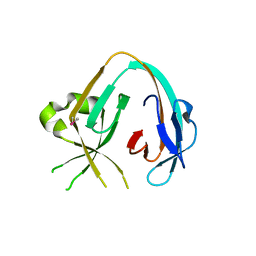 | | The Prp8 intein of Cryptococcus gattii | | Descriptor: | Pre-mRNA-processing-splicing factor 8 | | Authors: | Li, Z, Fu, B, Green, C.M, Lang, Y, Zhang, J, Oven, T.S, Li, X, Callahan, B.P, Chaturvedi, S, Belfort, M, Liao, G, Li, H. | | Deposit date: | 2018-10-30 | | Release date: | 2019-11-06 | | Last modified: | 2020-05-20 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Cisplatin protects mice from challenge ofCryptococcus neoformansby targeting the Prp8 intein.
Emerg Microbes Infect, 8, 2019
|
|
5UOC
 
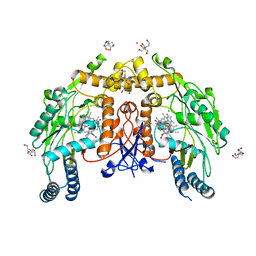 | | Structure of human endothelial nitric oxide synthase heme domain in complex with (S)-3-[(2-amino-4-methylquinolin-7-yl)methoxy]-5-(2-(methylamino)propyl)benzonitrile | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[(2-amino-4-methylquinolin-7-yl)methoxy]-5-[(2S)-2-(methylamino)propyl]benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2017-01-31 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nitrile in the Hole: Discovery of a Small Auxiliary Pocket in Neuronal Nitric Oxide Synthase Leading to the Development of Potent and Selective 2-Aminoquinoline Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UR9
 
 | | Enantiomer-Specific Binding of the Potent Antinociceptive Agent SBFI-26 to Anandamide transporters FABP5 | | Descriptor: | (1S,2S,3S,4S)-3-{[(naphthalen-1-yl)oxy]carbonyl}-2,4-diphenylcyclobutane-1-carboxylic acid, Fatty acid-binding protein, epidermal, ... | | Authors: | Hsu, H.-C, Li, H. | | Deposit date: | 2017-02-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.19800353 Å) | | Cite: | The Antinociceptive Agent SBFI-26 Binds to Anandamide Transporters FABP5 and FABP7 at Two Different Sites.
Biochemistry, 56, 2017
|
|
7E74
 
 | |
5UO9
 
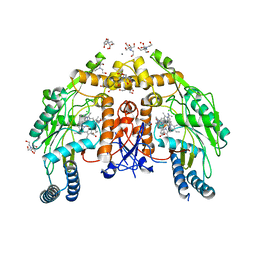 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 7-[(3-Ethyl-5-((methylamino)methyl)phenoxy)methyl]quinolin-2-amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 7-({3-ethyl-5-[(methylamino)methyl]phenoxy}methyl)quinolin-2-amine, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2017-01-31 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Nitrile in the Hole: Discovery of a Small Auxiliary Pocket in Neuronal Nitric Oxide Synthase Leading to the Development of Potent and Selective 2-Aminoquinoline Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UQP
 
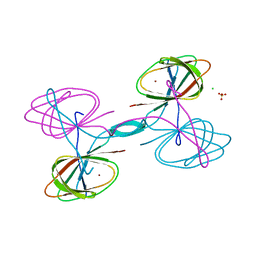 | | The crystal structure of cupin protein from Rhodococcus jostii RHA1 | | Descriptor: | CHLORIDE ION, Cupin, SULFATE ION, ... | | Authors: | Tan, K, Li, H, Clancy, S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-02-08 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of cupin protein from Rhodococcus jostii RHA1
To Be Published
|
|
5UO8
 
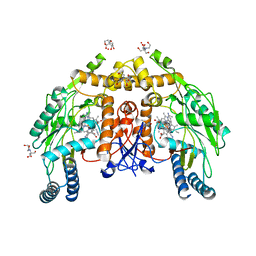 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 3-[(2-aminoquinolin-7-yl)methoxy]-5-((methylamino)methyl)benzonitrile | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[(2-aminoquinolin-7-yl)methoxy]-5-[(methylamino)methyl]benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2017-01-31 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Nitrile in the Hole: Discovery of a Small Auxiliary Pocket in Neuronal Nitric Oxide Synthase Leading to the Development of Potent and Selective 2-Aminoquinoline Inhibitors.
J. Med. Chem., 60, 2017
|
|
2M5Y
 
 | |
9ASH
 
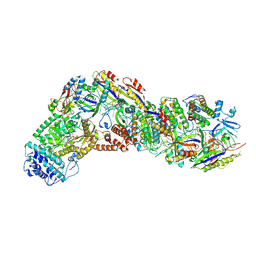 | | Cryo-EM structure of the active Lactococcus lactis Csm bound to target in post-cleavage stage | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR RNA, CRISPR system Cms endoribonuclease Csm3, ... | | Authors: | Wang, B, Goswami, H.N, Li, H. | | Deposit date: | 2024-02-25 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Molecular basis for cA6 synthesis by a type III-A CRISPR-Cas enzyme and its conversion to cA4 production.
Nucleic Acids Res., 52, 2024
|
|
9ASI
 
 | | Cryo-EM structure of the active Lactococcus lactis Csm bound to target in pre-cleavage stage | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CRISPR RNA, ... | | Authors: | Wang, B, Goswami, H.N, Li, H. | | Deposit date: | 2024-02-25 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Molecular basis for cA6 synthesis by a type III-A CRISPR-Cas enzyme and its conversion to cA4 production.
Nucleic Acids Res., 52, 2024
|
|
1Z0X
 
 | | Crystal structure of transcriptional regulator, tetR Family from Enterococcus faecalis V583 | | Descriptor: | CHLORIDE ION, transcriptional regulator, TetR family | | Authors: | Chang, C, Li, H, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-03-02 | | Release date: | 2005-04-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of transcriptional regulator, tetR Family from Enterococcus faecalis V583
To be Published
|
|
1Z6M
 
 | | Structure of Conserved Protein of Unknown Function from Enterococcus faecalis V583 | | Descriptor: | PHOSPHATE ION, conserved hypothetical protein | | Authors: | Nocek, B.P, Li, H, Collart, F, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-03-22 | | Release date: | 2005-05-03 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of a conserved hypothetical protein from Enterococcus faecalis V583
To be Published
|
|
