9IQF
 
 | |
9IQG
 
 | | Cryo-EM structure of MsRv1273c/72c from Mycobacterium smegmatis in the ATP|ADP+Vi-bound Occ (Vi) state | | Descriptor: | ABC transporter transmembrane region, ABC transporter, ATP-binding protein, ... | | Authors: | Lan, Y, Yu, J, Li, J. | | Deposit date: | 2024-07-12 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure and mechanism of a mycobacterial isoniazid efflux pump MsRv1273c/72c with a degenerate nucleotide-binding site.
Nat Commun, 16, 2025
|
|
2F6I
 
 | | Crystal structure of the ClpP protease catalytic domain from Plasmodium falciparum | | Descriptor: | ATP-dependent CLP protease, putative | | Authors: | Mulichak, A, Loppnau, P, Bray, J, Amani, M, Vedadi, M, Wasney, G, Finerty, P, Sundstrom, M, Weigelt, J, Edwards, A, Arrowsmith, C, Hui, R, Plotnikova, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-11-29 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The Clp chaperones and proteases of the human malaria parasite Plasmodium falciparum.
J.Mol.Biol., 404, 2010
|
|
9KWI
 
 | | Cryo-EM structure of MsRv1273c/72c(E553Q) mutant from Mycobacterium smegmatis in the ATP-bound Occ state | | Descriptor: | ABC transporter, ATP-binding protein, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Lan, Y, Yu, J, Li, J. | | Deposit date: | 2024-12-05 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structure and mechanism of a mycobacterial isoniazid efflux pump MsRv1273c/72c with a degenerate nucleotide-binding site.
Nat Commun, 16, 2025
|
|
2FBN
 
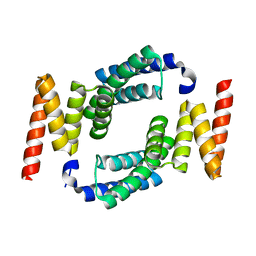 | | Plasmodium falciparum putative FK506-binding protein PFL2275c, C-terminal TPR-containing domain | | Descriptor: | 70 kDa peptidylprolyl isomerase, putative | | Authors: | Dong, A, Lew, J, Koeieradzki, I, Sundararajan, E, Melone, M, Wasney, G, Zhao, Y, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-12-09 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystallographic structure of the tetratricopeptide repeat domain of Plasmodium falciparum FKBP35 and its molecular interaction with Hsp90 C-terminal pentapeptide.
Protein Sci., 18, 2009
|
|
6IJX
 
 | | Crystal Structure of AKR1C1 complexed with meclofenamic acid | | Descriptor: | 2-[(2,6-dichloro-3-methyl-phenyl)amino]benzoic acid, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X, Zhao, Y, Zhang, L, Zhang, H, Chen, Y, Hu, X. | | Deposit date: | 2018-10-12 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Screening, synthesis, crystal structure, and molecular basis of 6-amino-4-phenyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitriles as novel AKR1C3 inhibitors.
Bioorg.Med.Chem., 26, 2018
|
|
3EFZ
 
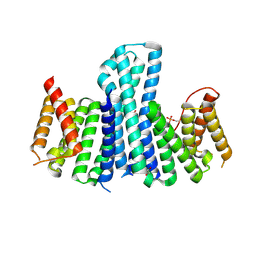 | | Crystal Structure of a 14-3-3 protein from cryptosporidium parvum (cgd1_2980) | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein | | Authors: | Wernimont, A.K, Dong, A, Qiu, W, Lew, J, Wasney, G.A, Vedadi, M, Kozieradzki, I, Zhao, Y, Ren, H, Alam, Z, Lin, Y.H, Sundstrom, M, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Hui, R, Brokx, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-10 | | Release date: | 2008-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Characterization of 14-3-3 proteins from Cryptosporidium parvum.
Plos One, 6, 2011
|
|
3EOZ
 
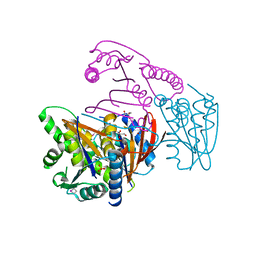 | | Crystal Structure of Phosphoglycerate Mutase from Plasmodium Falciparum, PFD0660w | | Descriptor: | GLYCEROL, PHOSPHATE ION, putative Phosphoglycerate mutase | | Authors: | Wernimont, A.K, Tempel, W, Lam, A, Zhao, Y, Lew, J, Lin, Y.H, Wasney, G, Vedadi, M, Kozieradzki, I, Cossar, D, Schapira, M, Weigelt, J, Arrowsmith, C.H, Bochkarev, A, Edwards, A.M, Hui, R, Pizarro, J, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization of a new phosphatase from Plasmodium.
Mol.Biochem.Parasitol., 179, 2011
|
|
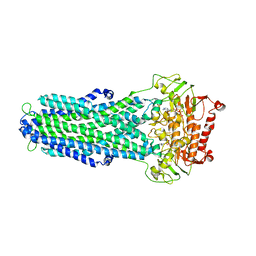
8IPT
 
 | |
8IPR
 
 | |
8IPS
 
 | |
8IPQ
 
 | |
9IM0
 
 | |
9ILY
 
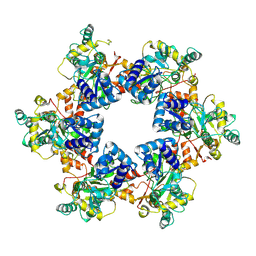 | |
9IM1
 
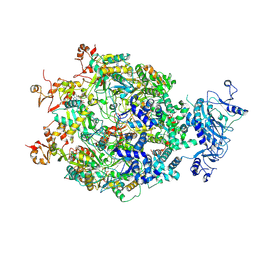 | | The Cryo-EM structure of MPXV E5 in complex with ssDNA in intermediate state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Cheng, Y.X, Han, P, Wang, H. | | Deposit date: | 2024-07-01 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Assembly and breakage of head-to-head double hexamer reveals mpox virus E5-catalyzed DNA unwinding initiation.
Nat Commun, 16, 2025
|
|
9IM2
 
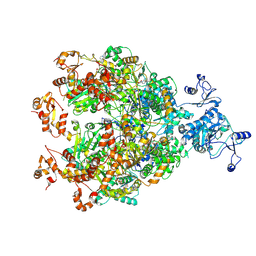 | | The Cryo-EM structure of MPXV E5 in complex with ssDNA in intermediate state 3 | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Cheng, Y.X, Han, P, Wang, H. | | Deposit date: | 2024-07-01 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Assembly and breakage of head-to-head double hexamer reveals mpox virus E5-catalyzed DNA unwinding initiation.
Nat Commun, 16, 2025
|
|
9ILZ
 
 | | The Cryo-EM structure of MPXV E5 in complex with ssDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Cheng, Y.X, Han, P, Wang, H. | | Deposit date: | 2024-07-01 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Assembly and breakage of head-to-head double hexamer reveals mpox virus E5-catalyzed DNA unwinding initiation.
Nat Commun, 16, 2025
|
|
9IM3
 
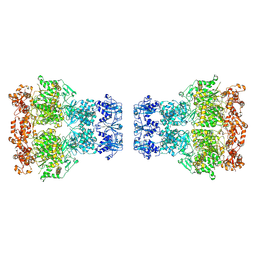 | |
5XGH
 
 | | Crystal structure of PI3K complex with an inhibitor | | Descriptor: | 3-[(4-fluorophenyl)methylamino]-5-(4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)phenol, GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Song, K, Yang, X, Zhao, Y, Jian, Z. | | Deposit date: | 2017-04-13 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | New Insights into PI3K Inhibitor Design using X-ray Structures of PI3K alpha Complexed with a Potent Lead Compound.
Sci Rep, 7, 2017
|
|
2QT5
 
 | | Crystal Structure of GRIP1 PDZ12 in Complex with the Fras1 Peptide | | Descriptor: | (ASN)(ASN)(LEU)(GLN)(ASP)(GLY)(THR)(GLU)(VAL), 1,2-ETHANEDIOL, ACETIC ACID, ... | | Authors: | Long, J, Wei, Z, Feng, W, Zhao, Y, Zhang, M. | | Deposit date: | 2007-08-01 | | Release date: | 2008-06-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Supramodular nature of GRIP1 revealed by the structure of its PDZ12 tandem in complex with the carboxyl tail of Fras1.
J.Mol.Biol., 375, 2008
|
|
8XXT
 
 | | ASFV RNAP M1249L C-tail occupied complex2 (MCOC2) | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Zhu, G.L, Zhu, Y, Zhu, Z.X, Sun, F, Zheng, H.X. | | Deposit date: | 2024-01-19 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of RNA polymerase complexes in African swine fever virus.
Nat Commun, 16, 2025
|
|
8XXP
 
 | | ASFV RNAP core complex | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Zhu, G.L, Zhu, Y, Zhu, Z.X, Sun, F, Zheng, H.X. | | Deposit date: | 2024-01-18 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of RNA polymerase complexes in African swine fever virus.
Nat Commun, 16, 2025
|
|
8XX4
 
 | | ASFV RNAP elongation complex | | Descriptor: | C122R, D339L, DNA (5'-D(P*CP*GP*CP*AP*AP*CP*GP*GP*CP*GP*A)-3'), ... | | Authors: | Zhu, G.L, Zhu, Y, Zhu, Z.X, Sun, F, Zheng, H.X. | | Deposit date: | 2024-01-17 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of RNA polymerase complexes in African swine fever virus.
Nat Commun, 16, 2025
|
|
8Y0E
 
 | | ASFV RNAP M1249L C-tail occupied complex4 (MCOC4) | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Zhu, G.L, Zhu, Y, Zhu, Z.X, Sun, F, Zheng, H.X. | | Deposit date: | 2024-01-22 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of RNA polymerase complexes in African swine fever virus.
Nat Commun, 16, 2025
|
|
8XX5
 
 | | ASFV RNAP M1249L C-tail occupied complex1 (MCOC1) | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Zhu, G.L, Zhu, Y, Zhu, Z.X, Sun, F, Zheng, H.X. | | Deposit date: | 2024-01-17 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis of RNA polymerase complexes in African swine fever virus.
Nat Commun, 16, 2025
|
|
