7BAK
 
 | |
7BAL
 
 | |
7X7W
 
 | | The X-ray Crystallographic Structure of D-Psicose 3-epimerase from Clostridia bacterium | | Descriptor: | D-PSICOSE 3-EPIMERASE, MANGANESE (II) ION | | Authors: | Li, Z.F, Ban, X.F, Xie, X.F, Tian, Y.X, Li, C.M, Gu, Z.B. | | Deposit date: | 2022-03-10 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal structure of a novel homodimeric D-allulose 3-epimerase from a Clostridia bacterium
Acta Crystallogr.,Sect.D, 78, 2022
|
|
7XB5
 
 | |
7CR7
 
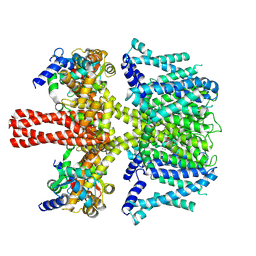 | | human KCNQ2-CaM in complex with retigabine | | Descriptor: | Calmodulin-3, Potassium voltage-gated channel subfamily KQT member 2, ethyl N-[2-azanyl-4-[(4-fluorophenyl)methylamino]phenyl]carbamate | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
7XS4
 
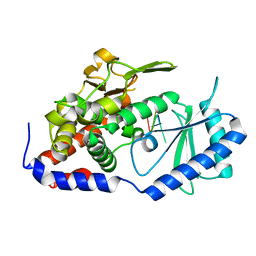 | | Crystal structure of URT1 in complex with AAAU RNA | | Descriptor: | RNA (5'-R(*AP*AP*AP*U)-3'), UTP:RNA uridylyltransferase 1 | | Authors: | Hu, Q, Zhu, L.R, lv, M.Q, Gong, Q.G. | | Deposit date: | 2022-05-12 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.846 Å) | | Cite: | Molecular mechanism underlying the di-uridylation activity of Arabidopsis TUTase URT1.
Nucleic Acids Res., 50, 2022
|
|
7CR0
 
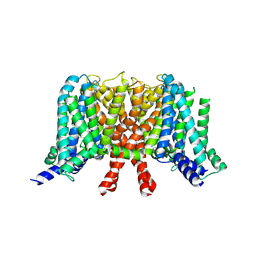 | | human KCNQ2 in apo state | | Descriptor: | Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
7CR3
 
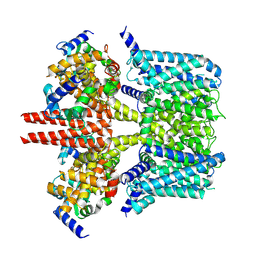 | | human KCNQ2-CaM in apo state | | Descriptor: | Calmodulin-3, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
7CR4
 
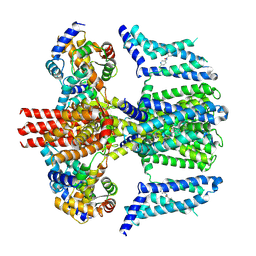 | | human KCNQ2-CaM in complex with ztz240 | | Descriptor: | Calmodulin-3, N-(6-chloranylpyridin-3-yl)-4-fluoranyl-benzamide, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
7FAI
 
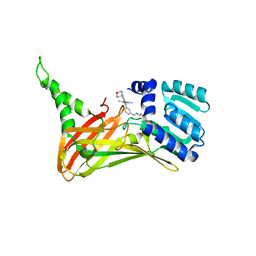 | | CARM1 bound with compound 9 | | Descriptor: | Histone-arginine methyltransferase CARM1, N'-[[3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)-5-methyl-6-(oxan-4-ylamino)pyrimidin-2-yl]phenyl]methyl]-N-methyl-ethane-1,2-diamine | | Authors: | Cao, D.Y, Li, J, Xiong, B. | | Deposit date: | 2021-07-06 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09749269 Å) | | Cite: | Structure-Based Discovery of Potent CARM1 Inhibitors for Solid Tumor and Cancer Immunology Therapy.
J.Med.Chem., 64, 2021
|
|
7FAJ
 
 | | CARM1 bound with compound 43 | | Descriptor: | Histone-arginine methyltransferase CARM1, N'-[[3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)-5-methyl-6-phenylazanyl-pyrimidin-2-yl]phenyl]methyl]-N-methyl-ethane-1,2-diamine | | Authors: | Cao, D.Y, Li, J, Xiong, B. | | Deposit date: | 2021-07-06 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2450726 Å) | | Cite: | Structure-Based Discovery of Potent CARM1 Inhibitors for Solid Tumor and Cancer Immunology Therapy.
J.Med.Chem., 64, 2021
|
|
9IY0
 
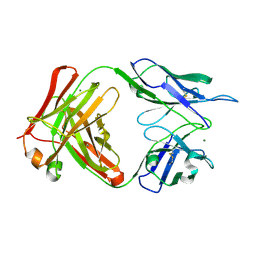 | | anti-HEV mAb 8H3 | | Descriptor: | Heavy chain of 8H3 Fab, Light chain of 8H3 Fab, MAGNESIUM ION | | Authors: | Minghua, Z, Lizhi, Z, Yang, H, Ying, G, Shaowei, L. | | Deposit date: | 2024-07-29 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis for the synergetic neutralization of hepatitis E virus by antibody-antibody interaction
To Be Published
|
|
6E2M
 
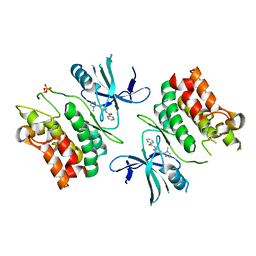 | | ASK1 kinase domain complex with inhibitor | | Descriptor: | Mitogen-activated protein kinase kinase kinase 5, N-[3-(4-methyl-4H-1,2,4-triazol-3-yl)phenyl]pyridine-2-carboxamide | | Authors: | Lansdon, E.B. | | Deposit date: | 2018-07-11 | | Release date: | 2018-09-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | ASK1 contributes to fibrosis and dysfunction in models of kidney disease.
J. Clin. Invest., 128, 2018
|
|
6E2N
 
 | | ASK1 kinase domain complex with inhibitor | | Descriptor: | Mitogen-activated protein kinase kinase kinase 5, N-[3-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)phenyl][3,4'-bipyridine]-2'-carboxamide | | Authors: | Lansdon, E.B. | | Deposit date: | 2018-07-11 | | Release date: | 2018-09-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | ASK1 contributes to fibrosis and dysfunction in models of kidney disease.
J. Clin. Invest., 128, 2018
|
|
6E2O
 
 | | ASK1 kinase domain complex with inhibitor | | Descriptor: | 4-(4-cyclopropyl-1H-imidazol-1-yl)-N-[3-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)phenyl]pyridine-2-carboxamide, Mitogen-activated protein kinase kinase kinase 5 | | Authors: | Lansdon, E.B. | | Deposit date: | 2018-07-11 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.389 Å) | | Cite: | ASK1 contributes to fibrosis and dysfunction in models of kidney disease.
J. Clin. Invest., 128, 2018
|
|
5XLL
 
 | | Dimer form of M. tuberculosis PknI sensor domain | | Descriptor: | Serine/threonine-protein kinase PknI | | Authors: | Rao, Z, Yan, Q. | | Deposit date: | 2017-05-10 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural Insight into the Activation of PknI Kinase from M. tuberculosis via Dimerization of the Extracellular Sensor Domain.
Structure, 25, 2017
|
|
5WQT
 
 | | Structure of a protein involved in pyroptosis | | Descriptor: | CITRIC ACID, GLYCEROL, Gasdermin-D | | Authors: | Kuang, S, Li, J. | | Deposit date: | 2016-11-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure insight of GSDMD reveals the basis of GSDMD autoinhibition in cell pyroptosis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XLM
 
 | | Monomer form of M.tuberculosis PknI sensor domain | | Descriptor: | Serine/threonine-protein kinase PknI | | Authors: | Rao, Z, Yan, Q. | | Deposit date: | 2017-05-10 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insight into the Activation of PknI Kinase from M. tuberculosis via Dimerization of the Extracellular Sensor Domain.
Structure, 25, 2017
|
|
5KH3
 
 | | Crystal structure of fragment (3-(5-Chloro-1,3-benzothiazol-2-yl)propanoic acid) bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(5-chloranyl-1,3-benzothiazol-2-yl)propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Dong, A, Ravichandran, M, Ferreira de Freitas, R, Schapira, M, Bountra, C, Edwards, A.M, Santhakumar, V, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin.
J. Med. Chem., 60, 2017
|
|
3I2N
 
 | | Crystal Structure of WD40 repeats protein WDR92 | | Descriptor: | WD repeat-containing protein 92 | | Authors: | Amaya, M.F, Li, Z, He, H, Seitova, A, Ni, S, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-29 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and function of WD40 domain proteins.
Protein Cell, 2, 2011
|
|
6CE6
 
 | | Structure of HDAC6 zinc-finger ubiquitin binding domain soaked with 3,3'-(benzo[1,2-d:5,4-d']bis(thiazole)-2,6-diyl)dipropionic acid | | Descriptor: | 3,3'-(benzo[1,2-d:5,4-d']bis[1,3]thiazole-2,6-diyl)dipropanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Ravichandran, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
6CED
 
 | | Crystal structure of fragment 3-(3-Methyl-4-oxo-3,4-dihydroquinazolin-2-yl)propanoic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(3-methyl-4-oxo-3,4-dihydroquinazolin-2-yl)propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Franzoni, I, Ravichandran, M, Lautens, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
3GFC
 
 | | Crystal Structure of Histone-binding protein RBBP4 | | Descriptor: | Histone-binding protein RBBP4 | | Authors: | Amaya, M.F, Dong, A, Li, Z, He, H, Ni, S, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-02-26 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of WD40 domain proteins.
Protein Cell, 2, 2011
|
|
6CE8
 
 | | Crystal structure of fragment 2-(Benzo[d]thiazol-2-yl)acetic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | (1,3-benzothiazol-2-yl)acetic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Ravichandran, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
6CEC
 
 | | Crystal structure of fragment 3-(3-Methoxy-2-quinoxalinyl)propanoic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(3-methoxyquinoxalin-2-yl)propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Franzoni, I, Ravichandran, M, Lautens, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
