8HVH
 
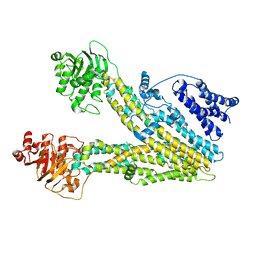 | | Cryo-EM structure of ABC transporter ABCC3 | | Descriptor: | ATP-binding cassette sub-family C member 3 | | Authors: | Wang, J, Wang, F.F, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2022-12-26 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Placing steroid hormones within the human ABCC3 transporter reveals a compatible amphiphilic substrate-binding pocket.
Embo J., 42, 2023
|
|
8HW2
 
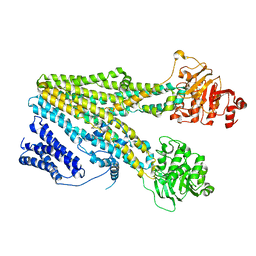 | | Cryo-EM structure of beta-estradiol 17-(beta-D-glucuronide)-bound human ABC transporter ABCC3 in nanodiscs | | Descriptor: | ATP-binding cassette sub-family C member 3, Estradiol-17beta-glucuronide | | Authors: | Wang, J, Wang, F.F, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2022-12-28 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Placing steroid hormones within the human ABCC3 transporter reveals a compatible amphiphilic substrate-binding pocket.
Embo J., 42, 2023
|
|
7W3T
 
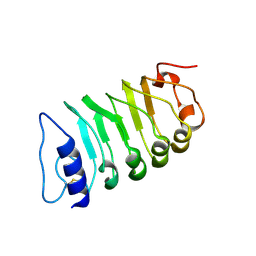 | | Cryo-EM structure of plant receptor like kinase NbBAK1 in RXEG1-BAK1-XEG1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Brassinosteroid insensitive 1-associated receptor kinase 1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Sun, Y, Wang, Y, Zhang, X.X, Chen, Z.D, Xia, Y.Q, Sun, Y.J, Zhang, M.M, Xiao, Y, Han, Z.F, Wang, Y.C, Chai, J.J. | | Deposit date: | 2021-11-26 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Plant receptor-like protein activation by a microbial glycoside hydrolase.
Nature, 610, 2022
|
|
7W3X
 
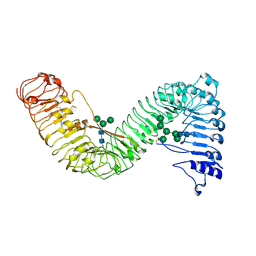 | | Cryo-EM structure of plant receptor like protein RXEG1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Membrane-localized LRR receptor-like protein, ... | | Authors: | Sun, Y, Wang, Y, Zhang, X.X, Chen, Z.D, Xia, Y.Q, Sun, Y.J, Zhang, M.M, Xiao, Y, Han, Z.F, Wang, Y.C, Chai, J.J. | | Deposit date: | 2021-11-26 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Plant receptor-like protein activation by a microbial glycoside hydrolase.
Nature, 610, 2022
|
|
7W3V
 
 | | Plant receptor like protein RXEG1 in complex with xyloglucanase XEG1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cell 12A endoglucanase, ... | | Authors: | Sun, Y, Wang, Y, Zhang, X.X, Chen, Z.D, Xia, Y.Q, Sun, Y.J, Zhang, M.M, Xiao, Y, Han, Z.F, Wang, Y.C, Chai, J.J. | | Deposit date: | 2021-11-26 | | Release date: | 2022-06-22 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Plant receptor-like protein activation by a microbial glycoside hydrolase.
Nature, 610, 2022
|
|
7WSF
 
 | |
7WSE
 
 | |
7WSH
 
 | | Cryo-EM structure of SARS-CoV-2 spike receptor-binding domain in complex with sea lion ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Li, S, Han, P, Qi, J. | | Deposit date: | 2022-01-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cross-species recognition and molecular basis of SARS-CoV-2 and SARS-CoV binding to ACE2s of marine animals.
Natl Sci Rev, 9, 2022
|
|
7WSG
 
 | |
4U3F
 
 | | Cytochrome bc1 complex from chicken with designed inhibitor bound | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CARDIOLIPIN, ... | | Authors: | Huang, L.-S, Zhu, X.-L, Yang, G.F, Berry, E.A. | | Deposit date: | 2014-07-21 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2312 Å) | | Cite: | Rational Design of Highly Potent and Slow-Binding Cytochrome bc1 Inhibitor as Fungicide by Computational Substitution Optimization
Sci Rep, 5, 2015
|
|
6BRB
 
 | | Novel non-antibody protein scaffold targeting CD40L | | Descriptor: | CD40 ligand, Tn3-like, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Oganesyan, V, Baca, M, Thisted, T, Grinberg, L, Wu, H, Dall'Acqua, W.F. | | Deposit date: | 2017-11-30 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | A CD40L-targeting protein reduces autoantibodies and improves disease activity in patients with autoimmunity.
Sci Transl Med, 11, 2019
|
|
4E1Z
 
 | |
3B9O
 
 | | long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN | | Descriptor: | Alkane monooxygenase, FLAVIN MONONUCLEOTIDE | | Authors: | Li, L, Yang, W, Xu, F, Bartlam, M, Rao, Z. | | Deposit date: | 2007-11-06 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN: unveiling the long-chain alkane hydroxylase
J.Mol.Biol., 376, 2008
|
|
3B9N
 
 | | Crystal structure of long-chain alkane monooxygenase (LadA) | | Descriptor: | Alkane monooxygenase | | Authors: | Li, L, Yang, W, Xu, F, Bartlam, M, Rao, Z. | | Deposit date: | 2007-11-06 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN: unveiling the long-chain alkane hydroxylase
J.Mol.Biol., 376, 2008
|
|
5XVW
 
 | |
6IS5
 
 | | P domain of GII.3-TV24 with A-tetrasaccharide complex | | Descriptor: | VP1 Capsid protein, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yang, Y. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural basis of host ligand specificity change of GII porcine noroviruses from their closely related GII human noroviruses.
Emerg Microbes Infect, 8, 2019
|
|
6IK4
 
 | | A Novel M23 Metalloprotease Pseudoalterin from Deep-sea | | Descriptor: | Elastinolytic metalloprotease, GLYCEROL, ZINC ION | | Authors: | Zhao, H.L, Tang, B.L, Yang, J, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2018-10-14 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A predator-prey interaction between a marine Pseudoalteromonas sp. and Gram-positive bacteria.
Nat Commun, 11, 2020
|
|
5Y53
 
 | |
5U5T
 
 | | Crystal structure of EED in complex with H3K27Me3 peptide and 3-(benzo[d][1,3]dioxol-4-ylmethyl)piperidine-1-carboximidamide | | Descriptor: | (3R)-3-[(2H-1,3-benzodioxol-4-yl)methyl]piperidine-1-carboximidamide, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED, ... | | Authors: | Bussiere, D, Shu, W. | | Deposit date: | 2016-12-07 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of EED Binders Allosterically Inhibiting the Epigenetic Polycomb Repressive Complex 2 (PRC2) Methyltransferase.
J. Med. Chem., 60, 2017
|
|
5U5K
 
 | |
8HML
 
 | |
7VWZ
 
 | | Cryo-EM structure of Rob-dependent transcription activation complex in a unique conformation | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Lin, W, Feng, Y, Shi, J. | | Deposit date: | 2021-11-12 | | Release date: | 2022-06-08 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of transcription activation by Rob, a pleiotropic AraC/XylS family regulator.
Nucleic Acids Res., 50, 2022
|
|
7VWY
 
 | |
6XQR
 
 | | OXA-48 bound by Compound 2.2 | | Descriptor: | Beta-lactamase, CHLORIDE ION, [1,1'-biphenyl]-4,4'-disulfonic acid | | Authors: | Taylor, D.M, Hu, L, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2020-07-10 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
7EFR
 
 | | Structure of SARS-CoV-2 spike receptor-binding domain in complex with high affinity ACE2 mutant (T27W,N330Y) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Lu, G.W, Ye, F, Lin, X. | | Deposit date: | 2021-03-23 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | S19W, T27W, and N330Y mutations in ACE2 enhance SARS-CoV-2 S-RBD binding toward both wild-type and antibody-resistant viruses and its molecular basis.
Signal Transduct Target Ther, 6, 2021
|
|
