7UA7
 
 | | Crystal structure of indoline 9 with KPC-2 | | Descriptor: | Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, tert-butyl {(3R)-3-[(1H-tetrazol-5-yl)carbamoyl]-1-[3-(trifluoromethyl)benzoyl]-2,3-dihydro-1H-indol-3-yl}carbamate | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2022-03-11 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | Probing the binding hot spots of KPC-2 carbapenemase using reversible tetrazole-based inhibitors
To Be Published
|
|
3HW5
 
 | | crystal structure of avian influenza virus PA_N in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Polymerase acidic protein | | Authors: | Zhao, C, Lou, Z, Guo, Y, Ma, M, Chen, Y, Liang, S, Rao, Z. | | Deposit date: | 2009-06-17 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Nucleoside monophosphate complex structures of the endonuclease domain from the influenza virus polymerase PA subunit reveal the substrate binding site inside the catalytic center
J.Virol., 83, 2009
|
|
3DW8
 
 | | Structure of a Protein Phosphatase 2A Holoenzyme with B55 subunit | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B alpha isoform, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Xu, Y, Chen, Y, Zhang, P, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2008-07-21 | | Release date: | 2008-10-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of a protein phosphatase 2A holoenzyme: insights into B55-mediated Tau dephosphorylation.
Mol.Cell, 31, 2008
|
|
1NYK
 
 | | Crystal Structure of the Rieske protein from Thermus thermophilus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, MAGNESIUM ION, Rieske iron-sulfur protein | | Authors: | Hunsicker-Wang, L.M, Heine, A, Chen, Y, Luna, E.P, Todaro, T, Zhang, Y.M, Williams, P.A, McRee, D.E, Hirst, J, Stout, C.D, Fee, J.A. | | Deposit date: | 2003-02-12 | | Release date: | 2003-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | High resolution structure of the soluble, respiratory-type Rieske protein from Thermus thermophilus: Analysis and Comparison
Biochemistry, 42, 2003
|
|
6MEY
 
 | | Crystal structure of KPC-2 with compound 9 | | Descriptor: | (2R)-2-phenyl-2-(phenylamino)-N-(1H-tetrazol-5-yl)acetamide, ACETATE ION, Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
3HW4
 
 | | Crystal structure of avian influenza A virus in complex with TMP | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein, THYMIDINE-5'-PHOSPHATE | | Authors: | Zhao, C, Lou, Z, Guo, Y, Ma, M, Chen, Y, Rao, Z. | | Deposit date: | 2009-06-17 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleoside monophosphate complex structures of the endonuclease domain from the influenza virus polymerase PA subunit reveal the substrate binding site inside the catalytic center
J.Virol., 83, 2009
|
|
6O3R
 
 | | Crystal Structure of NDM-1 D199N with Compound 7 | | Descriptor: | Carbapenem Hydrolyzing Class B Metallo beta lactamase NDM-1, ZINC ION, [(5-methoxy-7-methyl-2-oxo-2H-1-benzopyran-4-yl)methyl]phosphonic acid | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2019-02-27 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Heteroaryl Phosphonates as Noncovalent Inhibitors of Both Serine- and Metallocarbapenemases.
J.Med.Chem., 62, 2019
|
|
6NY7
 
 | | Crystal Structure of NDM-1 D199N with Compound 16 | | Descriptor: | Carbapenem Hydrolyzing Class B Metallo beta lactamase NDM-1, ZINC ION, [(5,7-dibromo-2-oxo-1,2-dihydroquinolin-4-yl)methyl]phosphonic acid | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2019-02-11 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Heteroaryl Phosphonates as Noncovalent Inhibitors of Both Serine- and Metallocarbapenemases.
J.Med.Chem., 62, 2019
|
|
7WQA
 
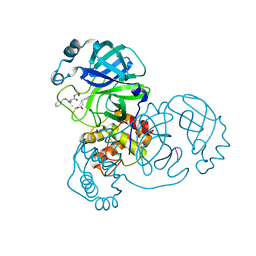 | | SARS-CoV-2 main protease in complex with Z-VAD-FMK | | Descriptor: | 3C-like proteinase, Z-VAD(OMe)-FMK | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chen, Y. | | Deposit date: | 2022-01-25 | | Release date: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SARS-CoV-2 main protease in complex with Z-VAD-FMK
To Be Published
|
|
5A6U
 
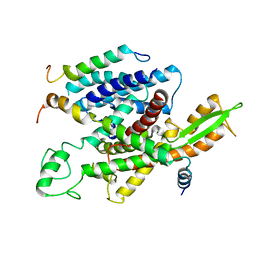 | | Native mammalian ribosome-bound Sec61 protein-conducting channel in the 'non-inserting' state | | Descriptor: | SEC61A, SEC61B, SEC61G | | Authors: | Pfeffer, S, Burbaum, L, Unverdorben, P, Pech, M, Chen, Y, Zimmermann, R, Beckmann, R, Foerster, F. | | Deposit date: | 2015-07-01 | | Release date: | 2015-10-07 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structure of the Native Sec61 Protein-Conducting Channel.
Nat.Commun., 6, 2015
|
|
6OOH
 
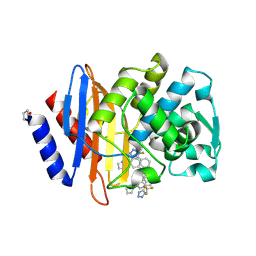 | | CTX-M-27 Beta Lactamase with Compound 14 | | Descriptor: | 3-(1H-pyrazol-1-yl)-N-[3-(1H-tetrazol-5-yl)phenyl]-5-(trifluoromethyl)benzamide, Beta-lactamase | | Authors: | Kemp, M, Chen, Y. | | Deposit date: | 2019-04-23 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | An Empirical Study of Amide-Heteroarene pi-Stacking Interactions Using Reversible Inhibitors of a Bacterial Serine Hydrolase.
Org Chem Front, 6, 2019
|
|
7WQ9
 
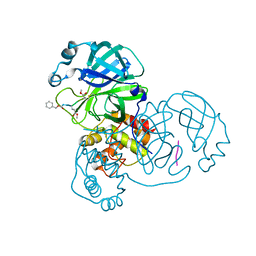 | | Crystal structure of SARS-CoV-2 main protease in complex with Z-IETD-FMK | | Descriptor: | 3C-like proteinase, Z-IETD-FMK | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chen, Y. | | Deposit date: | 2022-01-25 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease in complex with Z-IETD-FMK
To Be Published
|
|
7WQ8
 
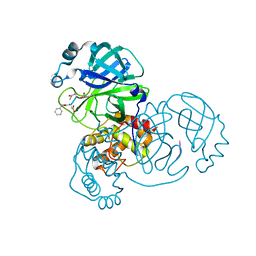 | | Crystal structure of SARS-CoV-2 main protease in complex with Z-DEVD-FMK | | Descriptor: | 3C-like proteinase, Z-DEVD-FMK | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chen, Y. | | Deposit date: | 2022-01-24 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease in complex with Z-DEVD-FMK
To Be Published
|
|
7WQK
 
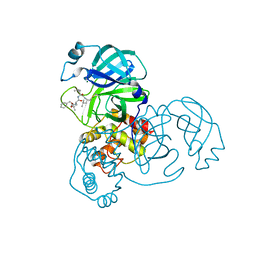 | | wild-type SARS-CoV-2 main protease in complex with MG-132 | | Descriptor: | 3C-like proteinase, MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chen, Y. | | Deposit date: | 2022-01-25 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | wild-type SARS-CoV-2 main protease in complex with MG-132
To Be Published
|
|
7WQB
 
 | | SARS-CoV-2 main protease mutant (P168A) in complex with MG-132 | | Descriptor: | 3C-like proteinase nsp5, MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chen, Y. | | Deposit date: | 2022-01-25 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | SARS-CoV-2 main protease mutant (P168A) in complex with MG-132
To Be Published
|
|
1Z6I
 
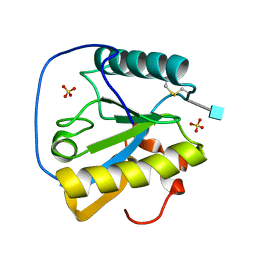 | | Crystal structure of the ectodomain of Drosophila transmembrane receptor PGRP-LCa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Peptidoglycan-recognition protein-LC, SULFATE ION | | Authors: | Chang, C.-I, Ihara, K, Chelliah, Y, Mengin-Lecreulx, D, Wakatsuki, S, Deisenhofer, J. | | Deposit date: | 2005-03-22 | | Release date: | 2005-07-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the ectodomain of Drosophila peptidoglycan-recognition protein LCa suggests a molecular mechanism for pattern recognition
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZZV
 
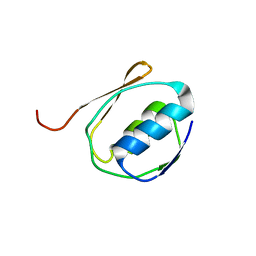 | | Solution NMR Structure of the Periplasmic Signaling Domain of the Outer Membrane Iron Transporter FecA from Escherichia coli. | | Descriptor: | Iron(III) dicitrate transport protein fecA | | Authors: | Ferguson, A.D, Amezcua, C.A, Chelliah, Y, Rosen, M.K, Deisenhofer, J. | | Deposit date: | 2005-06-14 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Signal transduction pathway of TonB-dependent transporters.
Proc.Natl.Acad.Sci.Usa, 2, 2006
|
|
1JJR
 
 | |
4PQG
 
 | | Crystal structure of the pneumococcal O-GlcNAc transferase GtfA in complex with UDP and GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycosyltransferase Gtf1, URIDINE-5'-DIPHOSPHATE | | Authors: | Shi, W.W, Jiang, Y.L, Zhu, F, Yang, Y.H, Wu, H, Ren, Y.M, Chen, Y, Zhou, C.Z. | | Deposit date: | 2014-03-03 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a Novel O-Linked N-Acetyl-d-glucosamine (O-GlcNAc) Transferase, GtfA, Reveals Insights into the Glycosylation of Pneumococcal Serine-rich Repeat Adhesins.
J.Biol.Chem., 289, 2014
|
|
6KMJ
 
 | | Crystal structure of Sth1 bromodomain in complex with H3K14Ac | | Descriptor: | GLYCEROL, Histone H3, Nuclear protein STH1/NPS1 | | Authors: | Chen, G, Li, W, Yan, F, Wang, D, Chen, Y. | | Deposit date: | 2019-07-31 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Structural Basis for Specific Recognition of H3K14 Acetylation by Sth1 in the RSC Chromatin Remodeling Complex.
Structure, 28, 2020
|
|
6KMB
 
 | | Crystal structure of Sth1 bromodomain | | Descriptor: | GLYCEROL, Nuclear protein STH1/NPS1 | | Authors: | Chen, G, Li, W, Yan, F, Wang, D, Chen, Y. | | Deposit date: | 2019-07-31 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structural Basis for Specific Recognition of H3K14 Acetylation by Sth1 in the RSC Chromatin Remodeling Complex.
Structure, 28, 2020
|
|
2A02
 
 | | Solution NMR Structure of the Periplasmic Signaling Domain of the Outer Membrane Iron Transporter PupA from Pseudomonas putida. | | Descriptor: | Ferric-pseudobactin 358 receptor | | Authors: | Ferguson, A.D, Amezcua, C.A, Chelliah, Y, Rosen, M.K, Deisenhofer, J. | | Deposit date: | 2005-06-15 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Signal transduction pathway of TonB-dependent transporters.
Proc.Natl.Acad.Sci.Usa, 2, 2006
|
|
4N9X
 
 | | Crystal Structure of the OCTAPRENYL-METHYL-METHOXY-BENZQ MOLECULE from Erwina carotovora subsp. atroseptica strain SCRI 1043 / ATCC BAA-672, Northeast Structural Genomics Consortium (NESG) Target EwR161 | | Descriptor: | Putative monooxygenase | | Authors: | Kuzin, A, Chen, Y, Lew, S, Seetharaman, J, Mao, L, Xiao, R, Owens, L.A, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-10-21 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Crystal Structure of the OCTAPRENYL-METHYL-METHOXY-BENZQ MOLECULE from Erwina carotovora subsp. atroseptica strain SCRI 1043 / ATCC BAA-672, Northeast Structural Genomics Consortium (NESG) Target EwR161
To be Published
|
|
2A4H
 
 | | Solution structure of Sep15 from Drosophila melanogaster | | Descriptor: | Selenoprotein Sep15 | | Authors: | Ferguson, A.D, Labunskyy, V.M, Fomenko, D.E, Chelliah, Y, Amezcua, C.A, Rizo, J, Gladyshev, V.N, Deisenhofer, J. | | Deposit date: | 2005-06-28 | | Release date: | 2005-12-13 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of the Selenoproteins Sep15 and SelM Reveal Redox Activity of a New Thioredoxin-like Family.
J.Biol.Chem., 281, 2006
|
|
4HRI
 
 | | Crystal structure of HetR in complex with a 21-bp palindromic DNA at the upstream of the hetP promoter from Anabaena | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*TP*GP*AP*GP*GP*GP*GP*TP*TP*AP*GP*AP*CP*CP*CP*CP*TP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*AP*GP*GP*GP*GP*TP*CP*TP*AP*AP*CP*CP*CP*CP*TP*CP*AP*T)-3'), ... | | Authors: | Hu, H.X, Jiang, Y.L, Zhao, M.X, Chen, Y, Zhang, C.C, Zhou, C.Z. | | Deposit date: | 2012-10-28 | | Release date: | 2013-04-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Structural and biochemical analyses of Anabaena HetR reveal insights into its binding to DNA targets and the inhibitory hexapeptide ERGSGR
To be Published
|
|
