8HFA
 
 | |
8HF9
 
 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | Chitin deacetylase, ZINC ION | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-10 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8J4Z
 
 | | Human 3-methylcrotonyl-CoA carboxylase in BCCP-CTS state with substrate | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | Authors: | Liu, D.S, Su, J.Y. | | Deposit date: | 2023-04-21 | | Release date: | 2024-04-24 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural insight into synergistic activation of human 3-methylcrotonyl-CoA carboxylase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
5UIX
 
 | |
6J2A
 
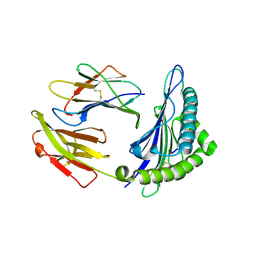 | | The structure of HLA-A*3003/NP44 | | Descriptor: | Beta-2-microglobulin, HLA-A*3003, NP44 | | Authors: | Zhu, S.Y, Liu, K.F, Chai, Y, Ding, C.M, Lv, J.X, Gao, F.G, Lou, Y.L, Liu, W.J. | | Deposit date: | 2018-12-31 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Divergent Peptide Presentations of HLA-A*30 Alleles Revealed by Structures With Pathogen Peptides.
Front Immunol, 10, 2019
|
|
6J1W
 
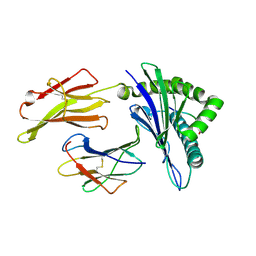 | | The structure of HLA-A*3001/RT313 | | Descriptor: | ALA-ILE-PHE-GLN-SER-SER-MET-THR-LYS, Beta-2-microglobulin, HLA-A*3001 | | Authors: | Zhu, S.Y, Liu, K.F, Chai, Y, Ding, C.M, Lv, J.X, Gao, G.F, Lou, Y.L, Liu, W.J. | | Deposit date: | 2018-12-29 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Divergent Peptide Presentations of HLA-A*30 Alleles Revealed by Structures With Pathogen Peptides.
Front Immunol, 10, 2019
|
|
8J7O
 
 | | Human pyruvate carboxylase in BCCP-CTS state without BC | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Pyruvate carboxylase, mitochondrial | | Authors: | Liu, D.S, Su, J.Y. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-08 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Structural insight into synergistic activation of human 3-methylcrotonyl-CoA carboxylase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8J73
 
 | | Methylcrotonoyl-CoA carboxylase core-Dimer | | Descriptor: | Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial | | Authors: | Liu, D.S, Su, J.Y. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-01 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structural insight into synergistic activation of human 3-methylcrotonyl-CoA carboxylase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8J78
 
 | | Human 3-methylcrotonyl-CoA carboxylase in BCCP-H2 state | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | Authors: | Liu, D.S, Su, J.Y. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-08 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Structural insight into synergistic activation of human 3-methylcrotonyl-CoA carboxylase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8J7D
 
 | | Human 3-methylcrotonyl-CoA carboxylase in BCCP-H1 state | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | Authors: | Liu, D.S, Su, J.Y. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-08 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insight into synergistic activation of human 3-methylcrotonyl-CoA carboxylase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
6J29
 
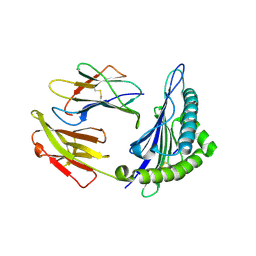 | | The structure of HLA-A*3003/MTB | | Descriptor: | Beta-2-microglobulin, HLA-A*3003, MTB | | Authors: | Zhu, S.Y, Liu, K.F, Chai, Y, Ding, C.M, Lv, J.X, Gao, F.G, Lou, Y.L, Liu, W.J. | | Deposit date: | 2018-12-31 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Divergent Peptide Presentations of HLA-A*30 Alleles Revealed by Structures With Pathogen Peptides.
Front Immunol, 10, 2019
|
|
8JAK
 
 | | Human MCC in MCCU state | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | Authors: | Liu, D.S, Su, J.Y, Tian, X.Y. | | Deposit date: | 2023-05-06 | | Release date: | 2024-05-15 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Structural insight into synergistic activation of human 3-methylcrotonyl-CoA carboxylase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8JAW
 
 | | Human MCC in MCCD state | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | Authors: | Liu, D.S, Su, J.Y, Tian, X.Y. | | Deposit date: | 2023-05-07 | | Release date: | 2024-05-15 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Structural insight into synergistic activation of human 3-methylcrotonyl-CoA carboxylase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8JXM
 
 | | Human 3-methylcrotonyl-CoA carboxylase in BCCP-H2 state with MCoA | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | Authors: | Liu, D.S, Su, J.Y, Tian, X.Y. | | Deposit date: | 2023-06-30 | | Release date: | 2024-07-03 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural insight into synergistic activation of human 3-methylcrotonyl-CoA carboxylase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8JXL
 
 | | Human 3-methylcrotonyl-CoA carboxylase in MCCU state with MCoA | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | Authors: | Liu, D.S, Su, J.Y, Tian, X.Y. | | Deposit date: | 2023-06-30 | | Release date: | 2024-07-03 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural insight into synergistic activation of human 3-methylcrotonyl-CoA carboxylase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8JXN
 
 | | Human 3-methylcrotonyl-CoA carboxylase in BCCP-H1 state with MCoA | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | Authors: | Liu, D.S, Su, J.Y, Tian, X.Y. | | Deposit date: | 2023-06-30 | | Release date: | 2024-07-03 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insight into synergistic activation of human 3-methylcrotonyl-CoA carboxylase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
6IEX
 
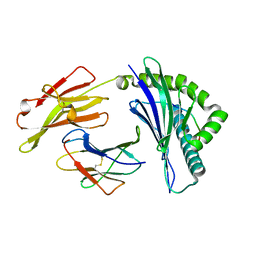 | | Crystal structure of HLA-B*4001 in complex with SARS-CoV derived peptide N216-225 GETALALLLL | | Descriptor: | Beta-2-microglobulin, GLY-GLU-THR-ALA-LEU-ALA-LEU-LEU-LEU-LEU, MHC class I antigen | | Authors: | Ji, W, Niu, L, Peng, W, Zhang, Y, Shi, Y, Qi, J, Gao, G.F, Liu, W.J. | | Deposit date: | 2018-09-17 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.314 Å) | | Cite: | Salt bridge-forming residues positioned over viral peptides presented by MHC class I impacts T-cell recognition in a binding-dependent manner.
Mol.Immunol., 112, 2019
|
|
6J1V
 
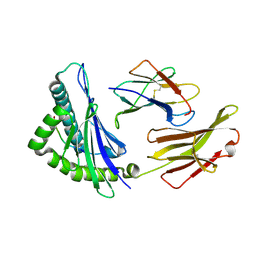 | | The structure of HLA-A*3003/RT313 | | Descriptor: | Beta-2-microglobulin, HLA-A*3003, RT313 | | Authors: | Zhu, S.Y, Liu, K.F, Chai, Y, Ding, C.M, Lv, J.X, Gao, G.F, Lou, Y.L, Liu, W.J. | | Deposit date: | 2018-12-29 | | Release date: | 2019-09-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Divergent Peptide Presentations of HLA-A*30 Alleles Revealed by Structures With Pathogen Peptides.
Front Immunol, 10, 2019
|
|
8K2V
 
 | | 3-Methylcrotonyl-CoA Carboxylase in MCCD state with Acetyl CoA | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ACETYL COENZYME *A, Methylcrotonoyl-CoA carboxylase beta chain, ... | | Authors: | Liu, D.S, Su, J.Y, Tian, X.Y. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-10 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural insight into synergistic activation of human 3-methylcrotonyl-CoA carboxylase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
5XOV
 
 | | Crystal structure of peptide-HLA-A24 bound to S19-2 V-delta/V-beta TCR | | Descriptor: | Beta-2-microglobulin, HIV-1 Nef138-10 peptide, HLA class I histocompatibility antigen, ... | | Authors: | Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2017-05-31 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.684 Å) | | Cite: | Conserved V delta 1 Binding Geometry in a Setting of Locus-Disparate pHLA Recognition by delta / alpha beta T Cell Receptors (TCRs): Insight into Recognition of HIV Peptides by TCRs.
J. Virol., 91, 2017
|
|
4MSV
 
 | | Crystal structure of FASL and DcR3 complex | | Descriptor: | GLYCEROL, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 6, ... | | Authors: | Liu, W, Ramagopal, U.A, Zhan, C, Bonanno, J.B, Bhosle, R.C, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-09-18 | | Release date: | 2013-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Complex of Human FasL and Its Decoy Receptor DcR3.
Structure, 24, 2016
|
|
4J6G
 
 | | CRYSTAL STRUCTURE OF LIGHT AND DcR3 COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Liu, W, Zhan, C, Bonanno, J.B, Bhosle, R.C, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-11 | | Release date: | 2013-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
4KG8
 
 | | Crystal structure of light mutant | | Descriptor: | Tumor necrosis factor ligand superfamily member 14 | | Authors: | Liu, W, Zhan, C, Kumar, P.R, Bonanno, J.B, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-04-28 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
7UV1
 
 | | Vicilin Ana o 1.0101 leader sequence residues 20-75 | | Descriptor: | Vicilin-like protein | | Authors: | Mueller, G.A, Foo, A.C.Y, DeRose, E.F. | | Deposit date: | 2022-04-29 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure and IgE Cross-Reactivity among Cashew, Pistachio, Walnut, and Peanut Vicilin-Buried Peptides.
J.Agric.Food Chem., 71, 2023
|
|
7UV4
 
 | | Pis v 3.0101 vicilin leader sequence residues 56-115 | | Descriptor: | Vicilin Pis v 3.0101 | | Authors: | Mueller, G.A, Foo, A.C.Y, DeRose, E.F. | | Deposit date: | 2022-04-29 | | Release date: | 2023-04-05 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structure and IgE Cross-Reactivity among Cashew, Pistachio, Walnut, and Peanut Vicilin-Buried Peptides.
J.Agric.Food Chem., 71, 2023
|
|
