3VM4
 
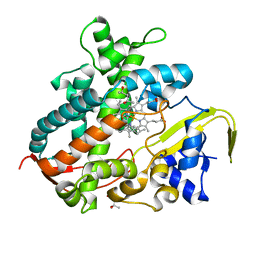 | | Cytochrome P450SP alpha (CYP152B1) in complex with (R)-ibuprophen | | Descriptor: | (2R)-2-[4-(2-methylpropyl)phenyl]propanoic acid, (4R)-2-METHYLPENTANE-2,4-DIOL, Fatty acid alpha-hydroxylase, ... | | Authors: | Fujishiro, T, Shoji, O, Nagano, S, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2011-12-08 | | Release date: | 2012-05-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Chiral-substrate-assisted stereoselective epoxidation catalyzed by H2O2-dependent cytochrome P450SP alpha
Chem Asian J, 7, 2012
|
|
7VPY
 
 | | Crystal structure of the neutralizing nanobody P86 against SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Nanobody, SULFATE ION | | Authors: | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
7VQ0
 
 | | Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing nanobodies P86 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
2RQL
 
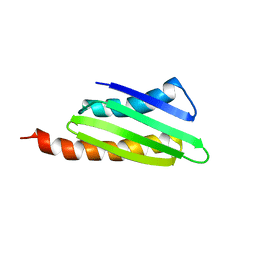 | |
2E4H
 
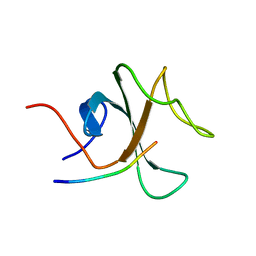 | |
2E3H
 
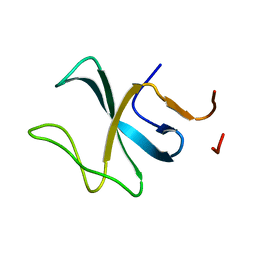 | |
2E3I
 
 | |
7W36
 
 | |
7DKJ
 
 | |
3WI2
 
 | | Crystal structure of PDE10A in complex with inhibitor | | Descriptor: | 2-({[1-phenyl-2-(propan-2-yl)-1H-benzimidazol-6-yl]oxy}methyl)quinoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Amano, Y. | | Deposit date: | 2013-09-04 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Design and synthesis of novel benzimidazole derivatives as phosphodiesterase 10A inhibitors with reduced CYP1A2 inhibition.
Bioorg.Med.Chem., 21, 2013
|
|
2D1V
 
 | | Crystal structure of DNA-binding domain of Bacillus subtilis YycF | | Descriptor: | Transcriptional regulatory protein yycF | | Authors: | Okajima, T, Okada, A, Watanabe, T, Yamamoto, K, Tanizawa, K, Utsumi, R. | | Deposit date: | 2005-09-01 | | Release date: | 2006-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Response regulator YycF essential for bacterial growth: X-ray crystal structure of the DNA-binding domain and its PhoB-like DNA recognition motif
Febs Lett., 582, 2008
|
|
3VZR
 
 | |
3VZQ
 
 | |
3VZP
 
 | | Crystal structure of PhaB from Ralstonia eutropha | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Acetoacetyl-CoA reductase, GLYCEROL, ... | | Authors: | Ikeda, K, Tanaka, Y, Tanaka, I, Yao, M. | | Deposit date: | 2012-10-15 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Directed evolution and structural analysis of NADPH-dependent Acetoacetyl Coenzyme A (Acetoacetyl-CoA) reductase from Ralstonia eutropha reveals two mutations responsible for enhanced kinetics
Appl.Environ.Microbiol., 79, 2013
|
|
3VZS
 
 | | Crystal structure of PhaB from Ralstonia eutropha in complex with Acetoacetyl-CoA and NADP | | Descriptor: | ACETOACETYL-COENZYME A, Acetoacetyl-CoA reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Ikeda, K, Tanaka, Y, Tanaka, I, Yao, M. | | Deposit date: | 2012-10-15 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Directed evolution and structural analysis of NADPH-dependent Acetoacetyl Coenzyme A (Acetoacetyl-CoA) reductase from Ralstonia eutropha reveals two mutations responsible for enhanced kinetics
Appl.Environ.Microbiol., 79, 2013
|
|
3W4J
 
 | | Crystal Structure of human DAAO in complex with coumpound 12 | | Descriptor: | 3-hydroxy-5-(2-phenylethyl)pyridin-2(1H)-one, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hondo, T, Warizaya, M, Niimi, T, Namatame, I, Yamaguchi, T, Nakanishi, K, Hamajima, T, Harada, K, Sakashita, H, Matsumoto, Y, Orita, M, Watanabe, T, Takeuchi, M. | | Deposit date: | 2013-01-09 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | 4-Hydroxypyridazin-3(2H)-one Derivatives as Novel d-Amino Acid Oxidase Inhibitors.
J.Med.Chem., 56, 2013
|
|
3W4K
 
 | | Crystal Structure of human DAAO in complex with coumpound 13 | | Descriptor: | 3-hydroxy-6-(2-phenylethyl)pyridazin-4(1H)-one, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hondo, T, Warizaya, M, Niimi, T, Namatame, I, Yamaguchi, T, Nakanishi, K, Hamajima, T, Harada, K, Sakashita, H, Matsumoto, Y, Orita, M, Watanabe, T, Takeuchi, M. | | Deposit date: | 2013-01-09 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | 4-Hydroxypyridazin-3(2H)-one Derivatives as Novel d-Amino Acid Oxidase Inhibitors.
J.Med.Chem., 56, 2013
|
|
3W4I
 
 | | Crystal Structure of human DAAO in complex with coumpound 8 | | Descriptor: | D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE, pyridine-2,3-diol | | Authors: | Hondo, T, Warizaya, M, Niimi, T, Namatame, I, Yamaguchi, T, Nakanishi, K, Hamajima, T, Harada, K, Sakashita, H, Matsumoto, Y, Orita, M, Watanabe, T, Takeuchi, M. | | Deposit date: | 2013-01-09 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 4-Hydroxypyridazin-3(2H)-one Derivatives as Novel d-Amino Acid Oxidase Inhibitors.
J.Med.Chem., 56, 2013
|
|
3AH4
 
 | | HA1 subcomponent of botulinum type C progenitor toxin complexed with galactose | | Descriptor: | Main hemagglutinin component, beta-D-galactopyranose | | Authors: | Nakamura, T, Tonozuka, T, Ide, A, Yuzawa, T, Oguma, K, Nishikawa, A. | | Deposit date: | 2010-04-13 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Sugar-binding sites of the HA1 subcomponent of Clostridium botulinum type C progenitor toxin
J.Mol.Biol., 376, 2008
|
|
3AH1
 
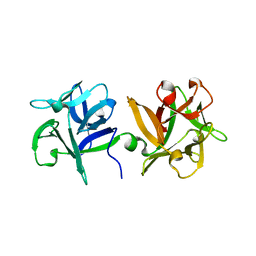 | | HA1 subcomponent of botulinum type C progenitor toxin complexed with N-acetylneuramic acid | | Descriptor: | Main hemagglutinin component, N-acetyl-beta-neuraminic acid | | Authors: | Nakamura, T, Tonozuka, T, Ide, A, Yuzawa, T, Oguma, K, Nishikawa, A. | | Deposit date: | 2010-04-13 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sugar-binding sites of the HA1 subcomponent of Clostridium botulinum type C progenitor toxin
J.Mol.Biol., 376, 2008
|
|
3AH2
 
 | | HA1 subcomponent of botulinum type C progenitor toxin complexed with N-acetylgalactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, Main hemagglutinin component | | Authors: | Nakamura, T, Tonozuka, T, Ide, A, Yuzawa, T, Oguma, K, Nishikawa, A. | | Deposit date: | 2010-04-13 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Sugar-binding sites of the HA1 subcomponent of Clostridium botulinum type C progenitor toxin
J.Mol.Biol., 376, 2008
|
|
