6ZFO
 
 | | Association of two complexes of largely structurally disordered Spike ectodomain with bound EY6A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, EY6A light chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-06-17 | | Release date: | 2020-07-08 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6Z97
 
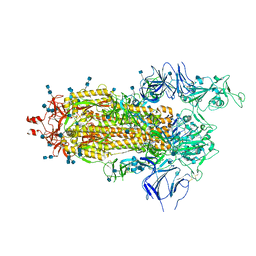 | | Structure of the prefusion SARS-CoV-2 spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Duyvesteyn, H.M.E, Ren, J, Zhao, Y, Zhou, D, Huo, J, Carrique, L, Malinauskas, T, Ruza, R.R, Shah, P.N.M, Fry, E.E, Owens, R, Stuart, D.I. | | Deposit date: | 2020-06-03 | | Release date: | 2020-07-01 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
6ZDH
 
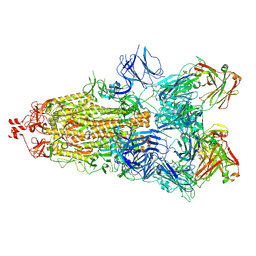 | | SARS-CoV-2 Spike glycoprotein in complex with a neutralizing antibody EY6A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-06-14 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8QZR
 
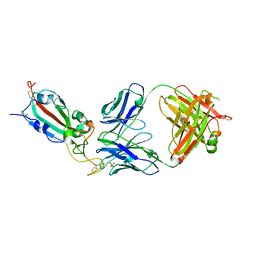 | | SARS-CoV-2 delta RBD complexed with BA.4/5-9 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BA.4/5-9 heavy chain, BA.4/5-9 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-10-29 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
5LRM
 
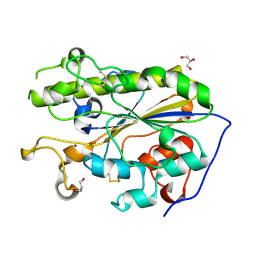 | | Structure of di-zinc MCR-1 in P41212 space group | | Descriptor: | GLYCEROL, ZINC ION, phosphatidylethanolamine transferase Mcr-1 | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2016-08-19 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into the Mechanistic Basis of Plasmid-Mediated Colistin Resistance from Crystal Structures of the Catalytic Domain of MCR-1.
Sci Rep, 7, 2017
|
|
4IDS
 
 | |
4IDM
 
 | |
4IHI
 
 | |
8C3V
 
 | | SARS-CoV-2 Delta-RBD complexed with BA.2-13 Fab and C1 nanobody | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, BA.2-13 heavy chain, BA.2-13 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-12-28 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Cell Rep, 42, 2023
|
|
8CBD
 
 | | SARS-CoV-2 Delta-RBD complexed with BA.4/5-1 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BA.4/5-1 heavy chain, BA.4/5-1 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
8CBF
 
 | | SARS-CoV-2 Delta-RBD complexed with Omi-42 and Beta-49 Fabs | | Descriptor: | Beta-49 heavy chain, Beta-49 light chain, CHLORIDE ION, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
8CBE
 
 | | SARS-CoV-2 Delta-RBD complexed with BA.4/5-2 and Beta-49 Fabs | | Descriptor: | BA.4/5-2 heavy chain, BA.4/5-2 light chain, Beta-49 heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
8CMA
 
 | | SARS-CoV-2 Delta-RBD complexed with BA.4/5-35 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BA.4/5-35 heavy chain, BA.4/5-35 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-02-18 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
8CMM
 
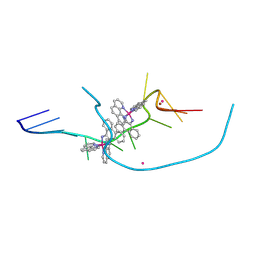 | | Re-pairing DNA - binding of a ruthenium phi complex to a double mismatch | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*AP*TP*AP*AP*TP*GP*CP*G)-3'), LITHIUM ION, POTASSIUM ION, ... | | Authors: | Prieto Otoya, T.D, Cardin, C.J, McQuaid, K.M. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Re-pairing DNA: binding of a ruthenium phi complex to a double mismatch.
Chem Sci, 15, 2024
|
|
8BBN
 
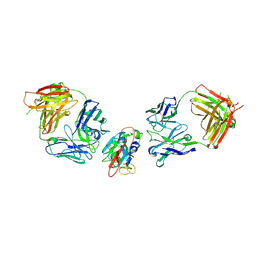 | | SARS-CoV-2 Delta-RBD complexed with BA.2-10 and EY6A Fabs | | Descriptor: | BA.2-10 heavy chain, BA.2-10 light chain, EY6A Heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-10-14 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Cell Rep, 42, 2023
|
|
8BBO
 
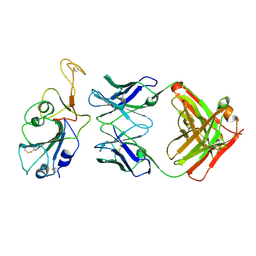 | | SARS-CoV-2 Delta-RBD complexed with BA.2-36 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IGH@ protein, Immunoglobulin kappa light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-10-14 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Cell Rep, 42, 2023
|
|
8BCZ
 
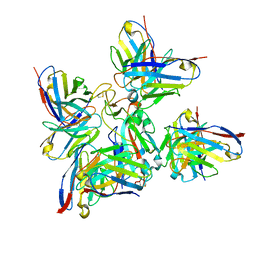 | | SARS-CoV-2 Delta-RBD complexed with Fabs BA.2-36, BA.2-23, EY6A and COVOX-45 | | Descriptor: | BA.2-23 heavy chain, BA.2-23 light chain, BA.2-36 heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2022-10-17 | | Release date: | 2023-03-22 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Cell Rep, 42, 2023
|
|
4JDC
 
 | |
7ZF8
 
 | | SARS-CoV-2 Omicron BA.2 RBD in complex with COVOX-150 Fab | | Descriptor: | COVOX-150 heavy chain, COVOX-150 light chain, Spike protein S1 | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZFE
 
 | | SARS-CoV-2 Omicron RBD in complex with Omi-32 Fab and nanobody C1 | | Descriptor: | Nanobody C1, Omi-32 heavy chain, Omi-32 light chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZFF
 
 | | Omi-42 Fab | | Descriptor: | GLYCEROL, Omi-42 Heavy chain, Omi-42 light chain | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZFD
 
 | | SARS-CoV-2 Omicron RBD in complex with Omi-25 Fab | | Descriptor: | Omi-25 heavy chain, Omi-25 light chain, Spike protein S1 | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZR9
 
 | | OMI-2 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE GLYCOPROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Omi-2 Fab heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2022-05-03 | | Release date: | 2022-06-01 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZFC
 
 | | SARS-CoV-2 Beta RBD in complex with nanobody C1, Omi-18 and Omi-31 Fabs | | Descriptor: | Nanobody C1, Omi-18 heavy chain, Omi-18 light chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZF6
 
 | | Omi-12 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, GLYCEROL, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
