7ZBE
 
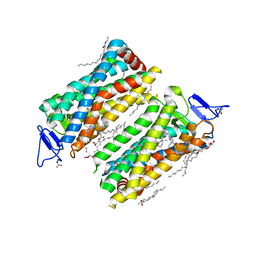 | | Dark state crystal structure of bovine rhodopsin in Lipidic Cubic Phase (SwissFEL) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gruhl, T, Weinert, T, Rodrigues, M.J, Milne, C, Ortolani, G, Nass, K, Nango, E, Sen, S, Johnson, P, Cirelli, C, Furrer, A, Mous, S, Skopintsev, P, James, D, Dworkowski, F, Baath, P, Kekilli, D, Oserov, D, Tanaka, R, Glover, H, Bacellar, C, Bruenle, S, Casadei, C, Diethelm, A, Gashi, D, Gotthard, G, Guixa-Gonzalez, R, Joti, Y, Kabanova, V, Knopp, G, Lesca, E, Ma, P, Martiel, I, Muehle, J, Owada, S, Pamula, F, Sarabi, D, Tejero, O, Tsai, C.J, Varma, N, Wach, A, Boutet, S, Tono, K, Nogly, P, Deupi, X, Iwata, S, Neutze, R, Standfuss, J, Schertler, G.F.X, Panneels, V. | | Deposit date: | 2022-03-23 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural changes direct the first molecular events of vision.
Nature, 615, 2023
|
|
7ZBC
 
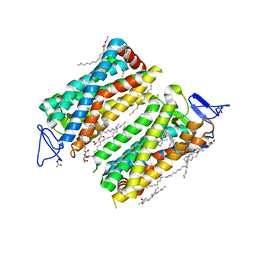 | | Dark state crystal structure of bovine rhodopsin in Lipidic Cubic Phase (SACLA) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gruhl, T, Weinert, T, Rodrigues, M.J, Milne, C, Ortolani, G, Nass, K, Nango, E, Sen, S, Johnson, P, Cirelli, C, Furrer, A, Mous, S, Skopintsev, P, James, D, Dworkowski, F, Baath, P, Kekilli, D, Oserov, D, Tanaka, R, Glover, H, Bacellar, C, Bruenle, S, Casadei, C, Diethelm, A, Gashi, D, Gotthard, G, Guixa-Gonzalez, R, Joti, Y, Kabanova, V, Knopp, G, Lesca, E, Ma, P, Martiel, I, Muehle, J, Owada, S, Pamula, F, Sarabi, S, Tejero, O, Tsai, C.J, Varma, N, Wach, A, Boutet, S, Tono, K, Nogly, P, Deupi, X, Iwata, S, Neutze, R, Standfuss, J, Schertler, G.F.X, Panneels, V. | | Deposit date: | 2022-03-23 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural changes direct the first molecular events of vision.
Nature, 615, 2023
|
|
6PRZ
 
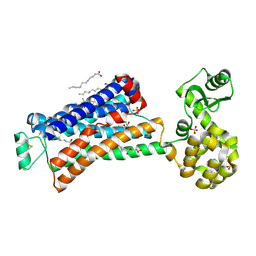 | | XFEL beta2 AR structure by ligand exchange from Alprenolol to Alprenolol. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-1-[(1-methylethyl)amino]-3-(2-prop-2-en-1-ylphenoxy)propan-2-ol, CHOLESTEROL, ... | | Authors: | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
6PS0
 
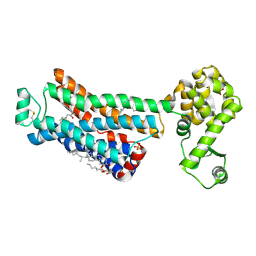 | | XFEL beta2 AR structure by ligand exchange from Alprenolol to Carazolol. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, CHOLESTEROL, ... | | Authors: | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
8A6E
 
 | | 100 picosecond light activated crystal structure of bovine rhodopsin in Lipidic Cubic Phase (SACLA) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gruhl, T, Weinert, T, Rodrigues, M.J, Milne, C.J, Ortolani, G, Nass, K, Nango, E, Sen, S, Johnson, P.J.M, Cirelli, C, Furrer, A, Mous, S, Skopintsev, P, James, D, Dworkowski, F, Baath, P, Kekilli, D, Oserov, D, Tanaka, R, Glover, H, Bacellar, C, Bruenle, S, Casadei, C.M, Diethelm, A.D, Gashi, D, Gotthard, G, Guixa-Gonzalez, R, Joti, Y, Kabanova, V, Knopp, G, Lesca, E, Ma, P, Martiel, I, Muehle, J, Owada, S, Pamula, F, Sarabi, D, Tejero, O, Tsai, C.J, Varma, N, Wach, A, Boutet, S, Tono, K, Nogly, P, Deupi, X, Iwata, S, Neutze, R, Standfuss, J, Schertler, G.F.X, Panneels, V. | | Deposit date: | 2022-06-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural changes direct the first molecular events of vision.
Nature, 615, 2023
|
|
8A6D
 
 | | 10 picosecond light activated crystal structure of bovine rhodopsin in Lipidic Cubic Phase | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gruhl, T, Weinert, T, Rodrigues, M.J, Milne, C.J, Ortolani, G, Nass, K, Nango, E, Sen, S, Johnson, P.J.M, Cirelli, C, Furrer, A, Mous, S, Skopintsev, P, James, D, Dworkowski, F, Baath, P, Kekilli, D, Oserov, D, Tanaka, R, Glover, H, Bacellar, C, Bruenle, S, Casadei, C.M, Diethelm, A.D, Gashi, D, Gotthard, G, Guixa-Gonzalez, R, Joti, Y, Kabanova, V, Knopp, G, Lesca, E, Ma, P, Martiel, I, Muehle, J, Owada, S, Pamula, F, Sarabi, D, Tejero, O, Tsai, C.J, Varma, N, Wach, A, Boutet, S, Tono, K, Nogly, P, Deupi, X, Iwata, S, Neutze, R, Standfuss, J, Schertler, G.F.X, Panneels, V. | | Deposit date: | 2022-06-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural changes direct the first molecular events of vision.
Nature, 615, 2023
|
|
8A6C
 
 | | 1 picosecond light activated crystal structure of bovine rhodopsin in Lipidic Cubic Phase | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gruhl, T, Weinert, T, Rodrigues, M.J, Milne, C.J, Ortolani, G, Nass, K, Nango, E, Sen, S, Johnson, P.J.M, Cirelli, C, Furrer, A, Mous, S, Skopintsev, P, James, D, Dworkowski, F, Baath, P, Kekilli, D, Oserov, D, Tanaka, R, Glover, H, Bacellar, C, Bruenle, S, Casadei, C.M, Diethelm, A.D, Gashi, D, Gotthard, G, Guixa-Gonzalez, R, Joti, Y, Kabanova, V, Knopp, G, Lesca, E, Ma, P, Martiel, I, Muehle, J, Owada, S, Pamula, F, Sarabi, D, Tejero, O, Tsai, C.J, Varma, N, Wach, A, Boutet, S, Tono, K, Nogly, P, Deupi, X, Iwata, S, Neutze, R, Standfuss, J, Schertler, G.F.X, Panneels, V. | | Deposit date: | 2022-06-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural changes direct the first molecular events of vision.
Nature, 615, 2023
|
|
6PS3
 
 | | XFEL beta2 AR structure by ligand exchange from Timolol to Carvedilol. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-1-(8H-CARBAZOL-4-YLOXY)-3-[2-(2-METHOXYPHENOXY)ETHYLAMINO]PROPAN-2-OL, CHOLESTEROL, ... | | Authors: | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
6PS6
 
 | | XFEL beta2 AR structure by ligand exchange from Timolol to Timolol. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-1-(tert-butylamino)-3-[(4-morpholin-4-yl-1,2,5-thiadiazol-3-yl)oxy]propan-2-ol, CHOLESTEROL, ... | | Authors: | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
6PS1
 
 | | XFEL beta2 AR structure by ligand exchange from Alprenolol to Timolol. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-1-(tert-butylamino)-3-[(4-morpholin-4-yl-1,2,5-thiadiazol-3-yl)oxy]propan-2-ol, CHOLESTEROL, ... | | Authors: | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
6PS8
 
 | | XFEL MT1R structure by ligand exchange from agomelatine to 2-phenylmelatonin. | | Descriptor: | Fusion protein of Melatonin receptor type 1A and GlgA glycogen synthase, N-[2-(5-methoxy-2-phenyl-1H-indol-3-yl)ethyl]acetamide | | Authors: | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
6PS4
 
 | | XFEL beta2 AR structure by ligand exchange from Timolol to ICI-118551. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S,3S)-1-[(7-methyl-2,3-dihydro-1H-inden-4-yl)oxy]-3-[(1-methylethyl)amino]butan-2-ol, CHOLESTEROL, ... | | Authors: | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
6PS5
 
 | | XFEL beta2 AR structure by ligand exchange from Timolol to Propranolol. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-(ISOPROPYLAMINO)-3-(1-NAPHTHYLOXY)-2-PROPANOL, CHOLESTEROL, ... | | Authors: | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
6PS2
 
 | | XFEL beta2 AR structure by ligand exchange from Timolol to Alprenolol. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-1-[(1-methylethyl)amino]-3-(2-prop-2-en-1-ylphenoxy)propan-2-ol, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, ... | | Authors: | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
6PS7
 
 | | XFEL A2aR structure by ligand exchange from LUF5843 to ZM241385. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
3WUN
 
 | |
5WS3
 
 | | Crystal structures of human orexin 2 receptor bound to the selective antagonist EMPA determined by serial femtosecond crystallography at SACLA | | Descriptor: | N-ethyl-2-[(6-methoxypyridin-3-yl)-(2-methylphenyl)sulfonyl-amino]-N-(pyridin-3-ylmethyl)ethanamide, OLEIC ACID, Orexin receptor type 2,GlgA glycogen synthase,Orexin receptor type 2, ... | | Authors: | Suno, R, Kimura, K, Nakane, T, Yamashita, K, Wang, J, Fujiwara, T, Yamanaka, Y, Im, D, Tsujimoto, H, Sasanuma, M, Horita, S, Hirokawa, T, Nango, E, Tono, K, Kameshima, T, Hatsui, T, Joti, Y, Yabashi, M, Shimamoto, K, Yamamoto, M, Rosenbaum, D.M, Iwata, S, Shimamura, T, Kobayashi, T. | | Deposit date: | 2016-12-05 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Human Orexin 2 Receptor Bound to the Subtype-Selective Antagonist EMPA.
Structure, 26, 2018
|
|
8AJZ
 
 | | Serial femtosecond crystallography structure of CO bound ba3- type cytochrome c oxidase at 2 milliseconds after irradiation by a 532 nm laser | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CARBON MONOXIDE, COPPER (II) ION, ... | | Authors: | Safari, C, Ghosh, S, Andersson, R, Johannesson, J, Donoso, A.V, Bath, P, Bosman, R, Dahl, P, Nango, E, Tanaka, R, Zoric, D, Svensson, E, Nakane, T, Iwata, S, Neutze, R, Branden, G. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial crystallography to track the dynamics of carbon monoxide in the active site of cytochrome c oxidase.
Sci Adv, 9, 2023
|
|
4YSD
 
 | | Room temperature structure of copper nitrite reductase from Geobacillus thermodenitrificans | | Descriptor: | ACETIC ACID, COPPER (II) ION, Nitrite reductase | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YSQ
 
 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 8.38 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YST
 
 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 24.9 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YSP
 
 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 8.32 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diedrichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YSO
 
 | | Copper nitrite reductase from Geobacillus thermodenitrificans - 0.064 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YSA
 
 | | Completely oxidized structure of copper nitrite reductase from Geobacillus thermodenitrificans | | Descriptor: | COPPER (II) ION, Nitrite reductase, SODIUM ION | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YSS
 
 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 16.7 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diedrichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
