2C76
 
 | | Functional Role of the Aromatic Cage in Human Monoamine Oxidase B: Structures and Catalytic Properties of Tyr435 Mutant Proteins | | Descriptor: | AMINE OXIDASE (FLAVIN-CONTAINING) B, FLAVIN-ADENINE DINUCLEOTIDE, N-PROPARGYL-1(S)-AMINOINDAN | | Authors: | Li, M, Binda, C, Mattevi, A, Edmondson, D.E. | | Deposit date: | 2005-11-17 | | Release date: | 2006-04-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional Role of the Aromatic Cage in Human Monoamine Oxidase B: Structures and Catalytic Properties of Tyr435 Mutant Proteins
Biochemistry, 45, 2006
|
|
2C73
 
 | | Functional Role of the Aromatic Cage in Human Monoamine Oxidase B: Structures and Catalytic Properties of Tyr435 Mutant Proteins | | Descriptor: | AMINE OXIDASE (FLAVIN-CONTAINING) B, FLAVIN-ADENINE DINUCLEOTIDE, N-PROPARGYL-1(S)-AMINOINDAN | | Authors: | Li, M, Binda, C, Mattevi, A, Edmondson, D.E. | | Deposit date: | 2005-11-17 | | Release date: | 2006-04-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional Role of the Aromatic Cage in Human Monoamine Oxidase B: Structures and Catalytic Properties of Tyr435 Mutant Proteins
Biochemistry, 45, 2006
|
|
2C70
 
 | | Functional Role of the Aromatic Cage in Human Monoamine Oxidase B: Structures and Catalytic Properties of Tyr435 Mutant Proteins | | Descriptor: | AMINE OXIDASE (FLAVIN-CONTAINING) B, FLAVIN-ADENINE DINUCLEOTIDE, P-NITRO-BENZYLAMINE | | Authors: | Li, M, Binda, C, Mattevi, A, Edmondson, D.E. | | Deposit date: | 2005-11-17 | | Release date: | 2006-04-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Functional Role of the Aromatic Cage in Human Monoamine Oxidase B: Structures and Catalytic Properties of Tyr435 Mutant Proteins
Biochemistry, 45, 2006
|
|
8W2R
 
 | | HIV-1 P5-IN intasome core | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (5'-D(*AP*CP*TP*GP*CP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*CP*CP*CP*G)-3'), DNA (5'-D(P*CP*GP*GP*GP*AP*AP*AP*AP*TP*CP*TP*CP*TP*AP*GP*CP*A)-3'), ... | | Authors: | Li, M, Craigie, R. | | Deposit date: | 2024-02-21 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | HIV-1 Integrase Assembles Multiple Species of Stable Synaptic Complex Intasomes That Are Active for Concerted DNA Integration In vitro.
J.Mol.Biol., 436, 2024
|
|
8W34
 
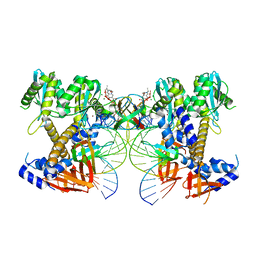 | | HIV-1 intasome core assembled with wild-type integrase, 1F | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (5'-D(*AP*CP*TP*GP*CP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*CP*CP*AP*C)-3'), DNA (5'-D(P*GP*TP*GP*GP*AP*AP*AP*AP*TP*CP*TP*CP*TP*AP*GP*CP*A)-3'), ... | | Authors: | Li, M, Craigie, R. | | Deposit date: | 2024-02-21 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | HIV-1 Integrase Assembles Multiple Species of Stable Synaptic Complex Intasomes That Are Active for Concerted DNA Integration In vitro.
J.Mol.Biol., 436, 2024
|
|
8W09
 
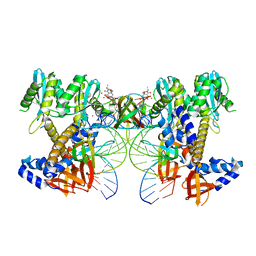 | | HIV-1 wild-type intasome core | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, Integrase, MAGNESIUM ION, ... | | Authors: | Li, M, Craigie, R. | | Deposit date: | 2024-02-13 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | HIV-1 Integrase Assembles Multiple Species of Stable Synaptic Complex Intasomes That Are Active for Concerted DNA Integration In vitro.
J.Mol.Biol., 436, 2024
|
|
2C72
 
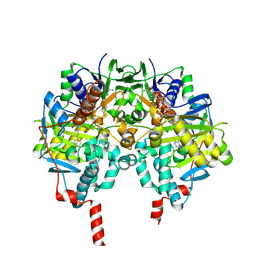 | | Functional Role of the Aromatic Cage in Human Monoamine Oxidase B: Structures and Catalytic Properties of Tyr435 Mutant Proteins | | Descriptor: | AMINE OXIDASE (FLAVIN-CONTAINING) B, FLAVIN-ADENINE DINUCLEOTIDE, N-PROPARGYL-1(S)-AMINOINDAN | | Authors: | Li, M, Binda, C, Mattevi, A, Edmondson, D.E. | | Deposit date: | 2005-11-17 | | Release date: | 2006-04-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional Role of the Aromatic Cage in Human Monoamine Oxidase B: Structures and Catalytic Properties of Tyr435 Mutant Proteins
Biochemistry, 45, 2006
|
|
2C75
 
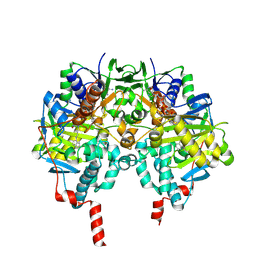 | | Functional Role of the Aromatic Cage in Human Monoamine Oxidase B: Structures and Catalytic Properties of Tyr435 Mutant Proteins | | Descriptor: | AMINE OXIDASE (FLAVIN-CONTAINING) B, FLAVIN-ADENINE DINUCLEOTIDE, N-PROPARGYL-1(S)-AMINOINDAN | | Authors: | Li, M, Binda, C, Mattevi, A, Edmondson, D.E. | | Deposit date: | 2005-11-17 | | Release date: | 2006-04-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional Role of the Aromatic Cage in Human Monoamine Oxidase B: Structures and Catalytic Properties of Tyr435 Mutant Proteins
Biochemistry, 45, 2006
|
|
2H2K
 
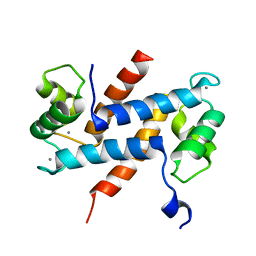 | | Crystal Structure Analysis of Human S100A13 | | Descriptor: | CALCIUM ION, Protein S100-A13 | | Authors: | Li, M, Zhang, P.F, Zhang, J.P, Chang, W.R. | | Deposit date: | 2006-05-19 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure study on human S100A13 at 2.0 A resolution
Biochem.Biophys.Res.Commun., 356, 2007
|
|
2JV5
 
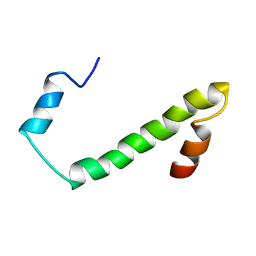 | | Nogo54 | | Descriptor: | Reticulon-4 | | Authors: | Li, M, Song, J. | | Deposit date: | 2007-09-12 | | Release date: | 2008-09-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure-based design of buffer-soluble Nogo54: a molecular mimic of Nogo-66 to inhibit CNS neuron regeneration
To be Published
|
|
2ANE
 
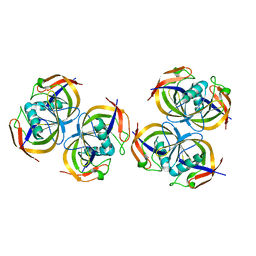 | | Crystal structure of N-terminal domain of E.Coli Lon Protease | | Descriptor: | ATP-dependent protease La | | Authors: | Li, M, Rasulova, F, Melnikov, E.E, Rotanova, T.V, Gustchina, A, Maurizi, M.R, Wlodawer, A. | | Deposit date: | 2005-08-11 | | Release date: | 2005-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of the N-terminal domain of E. coli Lon protease.
Protein Sci., 14, 2005
|
|
3SLZ
 
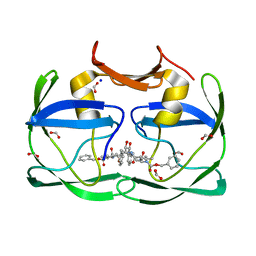 | | The crystal structure of XMRV protease complexed with TL-3 | | Descriptor: | FORMIC ACID, SODIUM ION, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate, ... | | Authors: | Li, M, Gustchina, A, Wlodawer, A. | | Deposit date: | 2011-06-27 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and biochemical characterization of the inhibitor complexes of xenotropic murine leukemia virus-related virus protease.
Febs J., 278, 2011
|
|
3SM2
 
 | | The crystal structure of XMRV protease complexed with Amprenavir | | Descriptor: | gag-pro-pol polyprotein, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | Authors: | Li, M, Gustchina, A, Wlodawer, A. | | Deposit date: | 2011-06-27 | | Release date: | 2011-10-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and biochemical characterization of the inhibitor complexes of xenotropic murine leukemia virus-related virus protease.
Febs J., 278, 2011
|
|
7VUL
 
 | |
6HB0
 
 | | Crystal structure of MSMEG_1712 from Mycobacterium smegmatis | | Descriptor: | ABC transporter periplasmic-binding protein YtfQ, ZINC ION | | Authors: | Li, M, Mueller, C, Einsle, O, Jessen-Trefzer, C. | | Deposit date: | 2018-08-09 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Detection and Characterization of a Mycobacterial L-Arabinofuranose ABC Transporter Identified with a Rapid Lipoproteomics Protocol.
Cell Chem Biol, 26, 2019
|
|
6HYH
 
 | | Crystal structure of MSMEG_1712 from Mycobacterium smegmatis in complex with Beta-D-Fucofuranose | | Descriptor: | Periplasmic binding protein/LacI transcriptional regulator, ZINC ION, beta-D-fucofuranose | | Authors: | Li, M, Mueller, C, Zhang, L, Einsle, O, Jessen-Trefzer, C. | | Deposit date: | 2018-10-21 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Detection and Characterization of a Mycobacterial L-Arabinofuranose ABC Transporter Identified with a Rapid Lipoproteomics Protocol.
Cell Chem Biol, 26, 2019
|
|
6JIX
 
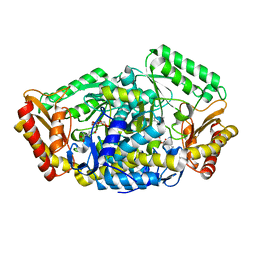 | | The cyrstal structure of taurine:2-oxoglutarate aminotransferase from Bifidobacterium kashiwanohense, in complex with PLP and glutamate | | Descriptor: | GLUTAMIC ACID, PYRIDOXAL-5'-PHOSPHATE, taurine:2-oxoglutarate aminotransferase | | Authors: | Li, M, Lin, L, Zhang, Y, Yuchi, Z. | | Deposit date: | 2019-02-23 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.647 Å) | | Cite: | Biochemical and structural investigation of taurine:2-oxoglutarate aminotransferase fromBifidobacterium kashiwanohense.
Biochem.J., 476, 2019
|
|
5X0X
 
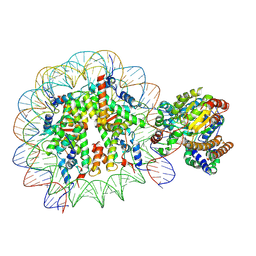 | | Complex of Snf2-Nucleosome complex with Snf2 bound to position +6 of the nucleosome | | Descriptor: | DNA (167-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, M, Liu, X, Xia, X, Chen, Z, Li, X. | | Deposit date: | 2017-01-23 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure.
Nature, 544, 2017
|
|
8J66
 
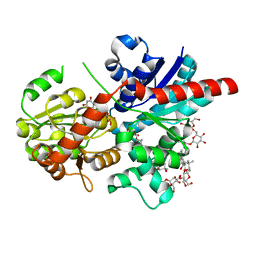 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with M3, state 2 | | Descriptor: | (2S,3S,4S,5R,6R)-2-(hydroxymethyl)-6-[[(2R,3S,4S,5R,6S)-6-[(3R,6S)-6-[(3S,8S,9R,10R,11S,13R,14S,17S)-3-[(2R,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-4,4,9,13,14-pentamethyl-11-oxidanyl-2,3,7,8,10,11,12,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-17-yl]-2-methyl-2-oxidanyl-heptan-3-yl]oxy-3,4,5-tris(oxidanyl)oxan-2-yl]methoxy]oxane-3,4,5-triol, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2023-04-24 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
8JW4
 
 | |
8JWI
 
 | |
8JWG
 
 | |
8JVH
 
 | |
8JWF
 
 | |
8FNP
 
 | | Structure of E138K/G140S/Q148H HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-28 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
