3IKJ
 
 | | Structural characterization for the nucleotide binding ability of subunit A mutant S238A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, V-type ATP synthase alpha chain | | Authors: | Kumar, A, Manimekali, M.S.S, Balakrishna, A.M, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2009-08-06 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nucleotide binding states of subunit A of the A-ATP synthase and the implication of P-loop switch in evolution.
J.Mol.Biol., 396, 2010
|
|
4R60
 
 | | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Proline dipeptidase, ... | | Authors: | Kumar, A, Ghosh, B, Are, V.N, Jamdar, S.N, Makde, R.D, Sharma, S.M. | | Deposit date: | 2014-08-22 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris
to be published
|
|
6JQV
 
 | | Crystal structure of Arabidopsis thaliana NRP2 | | Descriptor: | NAP1-related protein 2 | | Authors: | Kumar, A, Vasudevan, D. | | Deposit date: | 2019-04-01 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structural Characterization ofArabidopsis thalianaNAP1-Related Protein 2 (AtNRP2) and Comparison with its Homolog AtNRP1.
Molecules, 24, 2019
|
|
5C9L
 
 | | Crystal structure of native PLL lectin from Photorhabdus luminescens at 1.65 A resolution | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kumar, A, Sykorova, P, Demo, G, Dobes, P, Hyrsl, P, Wimmerova, M. | | Deposit date: | 2015-06-27 | | Release date: | 2016-10-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Novel Fucose-binding Lectin from Photorhabdus luminescens (PLL) with an Unusual Heptabladed beta-Propeller Tetrameric Structure.
J.Biol.Chem., 291, 2016
|
|
5CDL
 
 | | Proline dipeptidase from Deinococcus radiodurans (selenomethionine derivative) | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Proline dipeptidase | | Authors: | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R. | | Deposit date: | 2015-07-04 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proline dipeptidase from Deinococcus radiodurans (selenomethionine derivative)
To Be Published
|
|
5C9O
 
 | | Crystal structure of recombinant PLL lectin from Photorhabdus luminescens at 1.5 A resolution | | Descriptor: | GLYCEROL, PLL lectin | | Authors: | Kumar, A, Sykorova, P, Demo, G, Dobes, P, Hyrsl, P, Wimmerova, M. | | Deposit date: | 2015-06-28 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Novel Fucose-binding Lectin from Photorhabdus luminescens (PLL) with an Unusual Heptabladed beta-Propeller Tetrameric Structure.
J.Biol.Chem., 291, 2016
|
|
5CDV
 
 | | Proline dipeptidase from Deinococcus radiodurans R1 | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Proline dipeptidase, ... | | Authors: | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R. | | Deposit date: | 2015-07-05 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Proline dipeptidase from Deinococcus radiodurans R1 at 1.45 Angstrom resolution
To Be Published
|
|
5CIK
 
 | | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris in citrate condition | | Descriptor: | CITRIC ACID, GLYCEROL, Proline dipeptidase | | Authors: | Kumar, A, Are, V, Ghosh, B, Agrawal, U, Jamdar, S, Makde, R.D. | | Deposit date: | 2015-07-13 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris in citrate condition
To Be Published
|
|
5CDE
 
 | | R372A mutant of Xaa-Pro dipeptidase from Xanthomonas campestris | | Descriptor: | Proline dipeptidase, SULFATE ION, ZINC ION | | Authors: | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R. | | Deposit date: | 2015-07-03 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | R372A mutant of Xaa-Pro dipeptidase from Xanthomonas campestris at 1.85 Angstrom resolution
To Be Published
|
|
5C9P
 
 | | Crystal structure of recombinant PLL lectin complexed with L-fucose from Photorhabdus luminescens at 1.75 A resolution | | Descriptor: | GLYCEROL, PLL lectin, alpha-L-fucopyranose | | Authors: | Kumar, A, Sykorova, P, Demo, G, Dobes, P, Hyrsl, P, Wimmerova, M. | | Deposit date: | 2015-06-28 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Novel Fucose-binding Lectin from Photorhabdus luminescens (PLL) with an Unusual Heptabladed beta-Propeller Tetrameric Structure.
J.Biol.Chem., 291, 2016
|
|
6JMI
 
 | | Crystal structure of M.tuberculosis Rv0081 | | Descriptor: | SULFATE ION, Uncharacterized HTH-type transcriptional regulator Rv0081 | | Authors: | Kumar, A, Phulera, S, Mande, C.S. | | Deposit date: | 2019-03-11 | | Release date: | 2019-04-10 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | Structural basis of hypoxic gene regulation by the Rv0081 transcription factor of Mycobacterium tuberculosis.
Febs Lett., 593, 2019
|
|
7CXA
 
 | | Structure of human Galectin-3 CRD in complex with TD-139 belonging to P31 space group. | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, A. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Molecular mechanism of interspecies differences in the binding affinity of TD139 to Galectin-3.
Glycobiology, 31, 2021
|
|
7CXC
 
 | |
7CXB
 
 | | Structure of mouse Galectin-3 CRD in complex with TD-139 belonging to P6522 space group. | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, A. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Molecular mechanism of interspecies differences in the binding affinity of TD139 to Galectin-3.
Glycobiology, 31, 2021
|
|
7CXD
 
 | | Xray structure of rat Galectin-3 CRD in complex with TD-139 belonging to P121 space group | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, BROMIDE ION, Galectin-3 | | Authors: | Kumar, A. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Molecular mechanism of interspecies differences in the binding affinity of TD139 to Galectin-3.
Glycobiology, 31, 2021
|
|
7C9B
 
 | | Crystal structure of dipeptidase-E from Xenopus laevis | | Descriptor: | Alpha-aspartyl dipeptidase, CALCIUM ION, SODIUM ION | | Authors: | Kumar, A, Singh, R, Makde, R.D. | | Deposit date: | 2020-06-05 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of aspartyl dipeptidase from Xenopus laevis revealed ligand binding induced loop ordering and catalytic triad assembly.
Proteins, 90, 2022
|
|
8S77
 
 | | Soluble epoxide hydrolase in complex with PROTAC JSF234 | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional epoxide hydrolase 2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kumar, A, Schoenfeld, J, Hiesinger, K, Lillich, F, Proschak, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-02-29 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Soluble epoxide hydrolase in complex with PROTAC JSF234
To Be Published
|
|
8S75
 
 | | Soluble epoxide hydrolase in complex with PROTAC FL412 | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional epoxide hydrolase 2, TETRAETHYLENE GLYCOL, ... | | Authors: | Kumar, A, Schoenfeld, J, Hiesinger, K, Lillich, F, Proschak, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-02-29 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Soluble epoxide hydrolase in complex with PROTAC FL412
To Be Published
|
|
8S76
 
 | | Soluble epoxide hydrolase in complex with PROTAC JSF67 | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional epoxide hydrolase 2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kumar, A, Schoenfeld, J, Hiesinger, K, Lillich, F, Proschak, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-02-29 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Soluble epoxide hydrolase in complex with PROTAC JSF67
To Be Published
|
|
7FFP
 
 | | Crystal structure of di-peptidase-E from Xenopus laevis | | Descriptor: | ASPARTIC ACID, Alpha-aspartyl dipeptidase, CALCIUM ION | | Authors: | Kumar, A, Singh, R, Makde, R.D. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of aspartyl dipeptidase from Xenopus laevis revealed ligand binding induced loop ordering and catalytic triad assembly.
Proteins, 90, 2022
|
|
7GAN
 
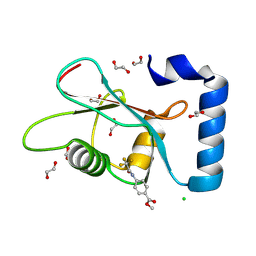 | | PanDDA analysis group deposition -- Crystal Structure of MAP1LC3B in complex with Z56767614 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Microtubule-associated proteins 1A/1B light chain 3B, ... | | Authors: | Kumar, A, Marples, P.G, Tomlinson, C.W.E, Fearon, D, von-Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-08-10 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7GAL
 
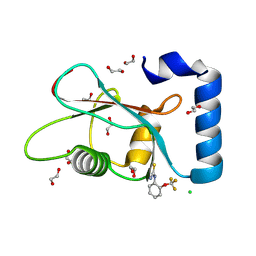 | | PanDDA analysis group deposition -- Crystal Structure of MAP1LC3B in complex with Z291279160 | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-(trifluoromethyloxy)phenyl]thiourea, CHLORIDE ION, ... | | Authors: | Kumar, A, Marples, P.G, Tomlinson, C.W.E, Fearon, D, von-Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-08-10 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7GAB
 
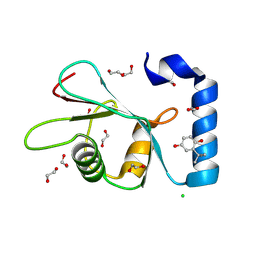 | | PanDDA analysis group deposition -- Crystal Structure of MAP1LC3B in complex with Z1255402624 | | Descriptor: | 1,2-ETHANEDIOL, 2-tert-butylbenzene-1,4-diol, CHLORIDE ION, ... | | Authors: | Kumar, A, Marples, P.G, Tomlinson, C.W.E, Fearon, D, von-Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-08-10 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7GAR
 
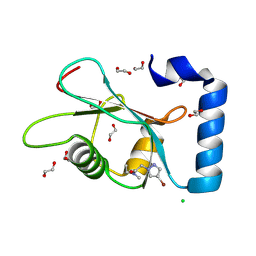 | | PanDDA analysis group deposition -- Crystal Structure of MAP1LC3B in complex with Z820676436 | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-bromanylpyrazol-1-yl)-~{N}-cyclopropyl-~{N}-methyl-ethanamide, CHLORIDE ION, ... | | Authors: | Kumar, A, Marples, P.G, Tomlinson, C.W.E, Fearon, D, von-Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-08-10 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7GAD
 
 | | PanDDA analysis group deposition -- Crystal Structure of MAP1LC3B in complex with Z1667545918 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Microtubule-associated proteins 1A/1B light chain 3B, ... | | Authors: | Kumar, A, Marples, P.G, Tomlinson, C.W.E, Fearon, D, von-Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-08-10 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
