6WZ4
 
 | | Complex of mutant (K173M) of Pseudomonas 7A Glutaminase-Asparaginase with L-Asp at pH 6. Covalent acyl-enzyme intermediate | | Descriptor: | ASPARTIC ACID, Glutaminase-asparaginase | | Authors: | Strzelczyk, P, Zhang, D, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2020-05-13 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Generalized enzymatic mechanism of catalysis by tetrameric L-asparaginases from mesophilic bacteria.
Sci Rep, 10, 2020
|
|
6WYY
 
 | | Crystal structure of Pseudomonas 7A Glutaminase-Asparaginase in complex with L-Glu at pH 6.5 | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Strzelczyk, P, Zhang, D, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2020-05-13 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Generalized enzymatic mechanism of catalysis by tetrameric L-asparaginases from mesophilic bacteria.
Sci Rep, 10, 2020
|
|
6WYZ
 
 | | Crystal structure of Pseudomonas 7A Glutaminase-Asparaginase (mutant K173M) in complex with D-Glu at pH 5.5 | | Descriptor: | D-GLUTAMIC ACID, Glutaminase-asparaginase | | Authors: | Strzelczyk, P, Zhang, D, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2020-05-13 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Generalized enzymatic mechanism of catalysis by tetrameric L-asparaginases from mesophilic bacteria.
Sci Rep, 10, 2020
|
|
8D1T
 
 | | Crystal structure of human USP30 in complex with a covalent inhibitor 552 and a Fab | | Descriptor: | (1R,2R,4S,7E)-7-[amino(sulfanyl)methylidene]-2-{[(1P)-3-chloro-3'-(1-cyanocyclopropyl)[1,1'-biphenyl]-4-carbonyl]amino}-7-azabicyclo[2.2.1]heptan-7-ium, 1,2-ETHANEDIOL, Ubiquitin carboxyl-terminal hydrolase 30, ... | | Authors: | Song, X, Butler, J, Li, C, Zhang, K, Zhang, D, Hao, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | TBD
To Be Published
|
|
2W0J
 
 | | Crystal structure of Chk2 in complex with NSC 109555, a specific inhibitor | | Descriptor: | 4,4'-DIACETYLDIPHENYLUREA-BIS(GUANYLHYDRAZONE), NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | Authors: | Lountos, G.T, Tropea, J.E, Zhang, D, Jobson, A.G, Pommier, Y, Shoemaker, R.H, Waugh, D.S. | | Deposit date: | 2008-08-18 | | Release date: | 2009-02-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of Checkpoint Kinase 2 in Complex with Nsc 109555, a Potent and Selective Inhibitor
Protein Sci., 18, 2009
|
|
7QHH
 
 | | Desensitized state of GluA1/2 AMPA receptor in complex with TARP-gamma 8 (TMD-LBD) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, GLUTAMIC ACID, ... | | Authors: | Herguedas, B, Kohegyi, B, Dohrke, J.N, Watson, J.F, Zhang, D, Ho, H, Shaikh, S, Lape, R, Krieger, J.M, Greger, I.H. | | Deposit date: | 2021-12-12 | | Release date: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanisms underlying TARP modulation of the GluA1/2-gamma 8 AMPA receptor.
Nat Commun, 13, 2022
|
|
7QHB
 
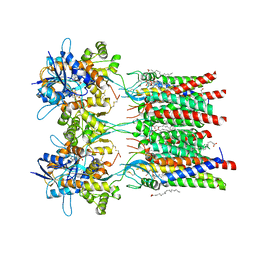 | | Active state of GluA1/2 in complex with TARP gamma 8, L-glutamate and CTZ | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, CYCLOTHIAZIDE, ... | | Authors: | Herguedas, B, Kohegyi, B, Zhang, D, Greger, I.H. | | Deposit date: | 2021-12-11 | | Release date: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanisms underlying TARP modulation of the GluA1/2-gamma 8 AMPA receptor.
Nat Commun, 13, 2022
|
|
6U0M
 
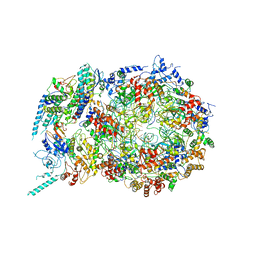 | | Structure of the S. cerevisiae replicative helicase CMG in complex with a forked DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA (15-MER), ... | | Authors: | Yuan, Z, Georgescu, R, Bai, L, Zhang, D, O'Donnell, M, Li, H. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | DNA unwinding mechanism of a eukaryotic replicative CMG helicase.
Nat Commun, 11, 2020
|
|
8HHK
 
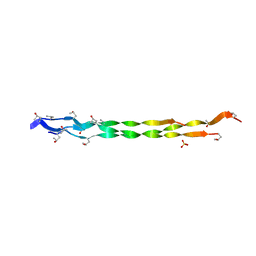 | |
8HHI
 
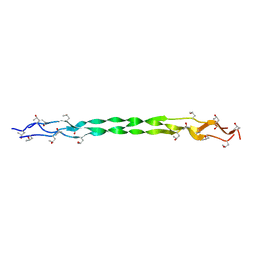 | |
6IS8
 
 | | Crystal structure of ZmMoc1 D115N mutant in complex with Holliday junction | | Descriptor: | DNA (33-MER), MAGNESIUM ION, Monokaryotic chloroplast 1, ... | | Authors: | Lin, Z, Lin, H, Zhang, D, Yuan, C. | | Deposit date: | 2018-11-15 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis of sequence-specific Holliday junction cleavage by MOC1.
Nat.Chem.Biol., 15, 2019
|
|
6IV5
 
 | | Crystal structure of arabidopsis N6-mAMP deaminase MAPDA | | Descriptor: | Adenosine/AMP deaminase family protein, PHOSPHATE ION, ZINC ION | | Authors: | Wu, B.X, Zhang, D, Nie, H.B, Shen, S.L, Li, S.S, Patel, D.J. | | Deposit date: | 2018-12-02 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Structure ofArabidopsis thaliana N6-methyl-AMP deaminase ADAL with bound GMP and IMP and implications forN6-methyl-AMP recognition and processing.
Rna Biol., 16, 2019
|
|
5ADY
 
 | | Cryo-EM structures of the 50S ribosome subunit bound with HflX | | Descriptor: | 23S RRNA, 50S RIBOSOMAL PROTEIN L1, 50S RIBOSOMAL PROTEIN L10, ... | | Authors: | Zhang, Y, Mandava, C.S, Cao, W, Li, X, Zhang, D, Li, N, Zhang, Y, Zhang, X, Qin, Y, Mi, K, Lei, J, Sanyal, S, Gao, N. | | Deposit date: | 2015-08-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Hflx is a Ribosome Splitting Factor Rescuing Stalled Ribosomes Under Stress Conditions
Nat.Struct.Mol.Biol., 22, 2015
|
|
3FCX
 
 | | Crystal structure of human esterase D | | Descriptor: | CALCIUM ION, MAGNESIUM ION, S-formylglutathione hydrolase | | Authors: | Wu, D, Li, Y, Song, G, Zhang, D, Shaw, N, Liu, Z.J. | | Deposit date: | 2008-11-24 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human esterase D: a potential genetic marker of retinoblastoma
Faseb J., 23, 2009
|
|
5OX6
 
 | | HIF prolyl hydroxylase 2 (PHD2/ EGLN1) in complex with Vadadustat | | Descriptor: | Egl nine homolog 1, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Chowdhury, R, Zhang, D, Schofield, C.J. | | Deposit date: | 2017-09-06 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular and cellular mechanisms of HIF prolyl hydroxylase inhibitors in clinical trials.
Chem Sci, 8, 2017
|
|
5OPC
 
 | | Factor Inhibiting HIF (FIH) in complex with zinc and Vadadustat | | Descriptor: | GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, SULFATE ION, ... | | Authors: | Leissing, T.M, Schofield, C.J, Clifton, I.J, Lu, X, Zhang, D. | | Deposit date: | 2017-08-09 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular and cellular mechanisms of HIF prolyl hydroxylase inhibitors in clinical trials.
Chem Sci, 8, 2017
|
|
5U8S
 
 | | Structure of eukaryotic CMG helicase at a replication fork | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA (26-MER), ... | | Authors: | Li, H, Li, B, Georgescu, R, Yuan, Z, Santos, R, Sun, J, Zhang, D, Yurieva, O, O'Donnell, M.E. | | Deposit date: | 2016-12-14 | | Release date: | 2017-01-25 | | Last modified: | 2020-01-01 | | Method: | ELECTRON MICROSCOPY (6.101 Å) | | Cite: | Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5U8T
 
 | | Structure of Eukaryotic CMG Helicase at a Replication Fork and Implications | | Descriptor: | Cell division control protein 45, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA replication complex GINS protein PSF1, ... | | Authors: | Li, B, Georgescu, R, Yuan, Z, Santos, R, Sun, J, Zhang, D, Yurieva, O, Li, H, O'Donnell, M.E. | | Deposit date: | 2016-12-15 | | Release date: | 2017-02-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1U6D
 
 | |
1DTH
 
 | | METALLOPROTEASE | | Descriptor: | 4-(N-HYDROXYAMINO)-2R-ISOBUTYL-2S-(2-THIENYLTHIOMETHYL)SUCCINYL-L-PHENYLALANINE-N-METHYLAMIDE, ATROLYSIN C, CALCIUM ION, ... | | Authors: | Botos, I, Scapozza, L, Zhang, D, Liotta, L.A, Meyer, E.F. | | Deposit date: | 1996-02-12 | | Release date: | 1997-02-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Batimastat, a potent matrix mealloproteinase inhibitor, exhibits an unexpected mode of binding.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
6MEP
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC3437 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Tyrosine-protein kinase Mer, ... | | Authors: | Da, C, Zhang, D, Stashko, M.A, Cheng, A, Hunter, D, Norris-Drouin, J, Graves, L, Machius, M, Miley, M.J, DeRyckere, D, Earp, H.S, Graham, D.K, Frye, S.V, Wang, X, Kireev, D. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.893 Å) | | Cite: | Data-Driven Construction of Antitumor Agents with Controlled Polypharmacology.
J.Am.Chem.Soc., 141, 2019
|
|
6M24
 
 | | Uncommon structural features of rabbit MHC class I (RLA-A1) complexed with rabbit haemorrhagic disease virus (RHDV) derived peptide, VP60-2 | | Descriptor: | Beta-2-microglobulin, RLA class I histocompatibility antigen, alpha chain 19-1, ... | | Authors: | Zhang, Q.X, Liu, K.F, Yue, C, Zhang, D, Lu, D, Xiao, W.L, Liu, P.P, Zhao, Y.Z, Gao, G.L, Ding, C.M, Lyu, J.X, Liu, W.J. | | Deposit date: | 2020-02-26 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Strict Assembly Restriction of Peptides from Rabbit Hemorrhagic Disease Virus Presented by Rabbit Major Histocompatibility Complex Class I Molecule RLA-A1.
J.Virol., 94, 2020
|
|
6M2J
 
 | | Uncommon structural features of rabbit MHC class I (RLA-A1) complexed with rabbit haemorrhagic disease virus (RHDV) derived peptide, VP60-1 | | Descriptor: | Beta-2-microglobulin, RLA class I histocompatibility antigen, alpha chain 19-1, ... | | Authors: | Zhang, Q.X, Liu, K.F, Yue, C, Zhang, D, Lu, D, Xiao, W.L, Liu, P.P, Zhao, Y.Z, Gao, G.L, Ding, C.M, Lyu, J.X, Liu, W.J. | | Deposit date: | 2020-02-27 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Strict Assembly Restriction of Peptides from Rabbit Hemorrhagic Disease Virus Presented by Rabbit Major Histocompatibility Complex Class I Molecule RLA-A1.
J.Virol., 94, 2020
|
|
6M2K
 
 | | Uncommon structural features of rabbit MHC class I (RLA-A1) complexed with rabbit haemorrhagic disease virus (RHDV) derived peptide, VP60-10 | | Descriptor: | Beta-2-microglobulin, RLA class I histocompatibility antigen, alpha chain 19-1, ... | | Authors: | Zhang, Q.X, Liu, K.F, Yue, C, Zhang, D, Lu, D, Xiao, W.L, Liu, P.P, Zhao, Y.Z, Gao, G.L, Ding, C.M, Lyu, J.X, Liu, W.J. | | Deposit date: | 2020-02-27 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Strict Assembly Restriction of Peptides from Rabbit Hemorrhagic Disease Virus Presented by Rabbit Major Histocompatibility Complex Class I Molecule RLA-A1.
J.Virol., 94, 2020
|
|
6K7T
 
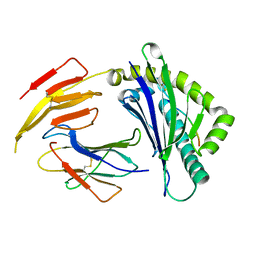 | | Crystal structure of bat (Pteropus Alecto) MHC class I Ptal-N*01:01 in complex with Hendra virus-derived peptide HeV1--human beta-2 microglobulin | | Descriptor: | Beta-2-microglobulin, HeV1, MHC class I antigen | | Authors: | Lu, D, Liu, K.F, Zhang, D, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, F.G, Liu, W.J. | | Deposit date: | 2019-06-08 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
