6E7D
 
 | | Structure of the inhibitory NKR-P1B receptor bound to the host-encoded ligand, Clr-b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain family 2 member D, Killer cell lectin-like receptor subfamily B member 1B allele B, ... | | Authors: | Balaji, G.R, Rossjohn, J, Berry, R. | | Deposit date: | 2018-07-26 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Recognition of host Clr-b by the inhibitory NKR-P1B receptor provides a basis for missing-self recognition.
Nat Commun, 9, 2018
|
|
4DUM
 
 | | Co-crystal structure of eIF4E with inhibitor | | Descriptor: | (4-{7-[2-(4-chlorophenoxy)ethyl]-2-(methylamino)-6-oxo-6,7-dihydro-1H-purin-8-yl}phenyl)phosphonic acid, 1,2-ETHANEDIOL, Eukaryotic translation initiation factor 4E | | Authors: | Min, X, Johnstone, S, Walker, N, Wang, Z. | | Deposit date: | 2012-02-22 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure-Guided Design, Synthesis, and Evaluation of Guanine-Derived Inhibitors of the eIF4E mRNA-Cap Interaction.
J.Med.Chem., 55, 2012
|
|
2FXT
 
 | | Crystal Structure of Yeast Tim44 | | Descriptor: | Import inner membrane translocase subunit TIM44 | | Authors: | Josyula, R, Sha, B. | | Deposit date: | 2006-02-06 | | Release date: | 2007-02-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of Yeast Mitochondrial Peripheral Membrane Protein Tim44p C-terminal Domain.
J.Mol.Biol., 359, 2006
|
|
1XAO
 
 | | Hsp40-Ydj1 dimerization domain | | Descriptor: | Mitochondrial protein import protein MAS5 | | Authors: | Wu, Y, Sha, B. | | Deposit date: | 2004-08-26 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The crystal structure of the C-terminal fragment of yeast Hsp40 Ydj1 reveals novel dimerization motif for Hsp40
J.Mol.Biol., 346, 2005
|
|
1YDN
 
 | | Crystal Structure of the HMG-CoA Lyase from Brucella melitensis, Northeast Structural Genomics Target LR35. | | Descriptor: | CALCIUM ION, HYDROXYMETHYLGLUTARYL-COA LYASE | | Authors: | Forouhar, F, Abashidze, M, Hussain, M, Vorobiev, S.M, Xiao, R, Ciano, M, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-12-24 | | Release date: | 2005-07-05 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of two bacterial 3-hydroxy-3-methylglutaryl-CoA lyases suggest a common catalytic mechanism among a family of TIM barrel metalloenzymes cleaving carbon-carbon bonds.
J.Biol.Chem., 281, 2006
|
|
1YDO
 
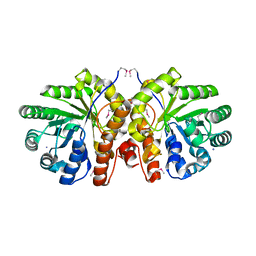 | | Crystal Structure of the Bacillis subtilis HMG-CoA Lyase, Northeast Structural Genomics Target SR181. | | Descriptor: | HMG-CoA Lyase, IODIDE ION | | Authors: | Forouhar, F, Hussain, M, Edstrom, W, Vorobiev, S.M, Xiao, R, Ciano, M, Shih, L, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-12-24 | | Release date: | 2005-07-05 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structures of two bacterial 3-hydroxy-3-methylglutaryl-CoA lyases suggest a common catalytic mechanism among a family of TIM barrel metalloenzymes cleaving carbon-carbon bonds.
J.Biol.Chem., 281, 2006
|
|
9C49
 
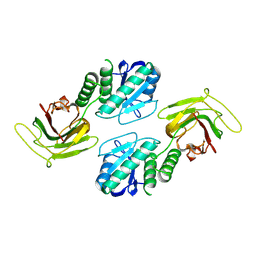 | |
7NOW
 
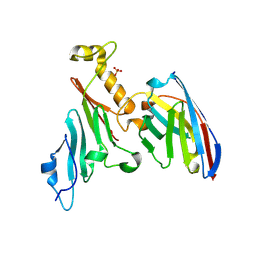 | |
7NQA
 
 | | Crystal structure of Nucleoporin-98 nanobody MS98-6 complex solved at 2.2A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Anti-Nup98 Nanobody MS98-6, Nuclear pore complex protein Nup98-Nup96, ... | | Authors: | Sola-Colom, M, Trakhanov, S, Goerlich, D. | | Deposit date: | 2021-03-01 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A checkpoint function for Nup98 in nuclear pore formation suggested by novel inhibitory nanobodies.
Embo J., 2024
|
|
5TZN
 
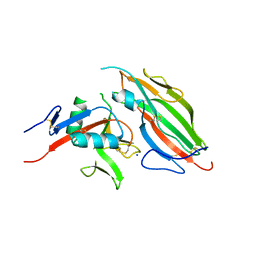 | | Structure of the viral immunoevasin m12 (Smith) bound to the natural killer cell receptor NKR-P1B (B6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein family protein m12, ... | | Authors: | Berry, R, Rossjohn, J. | | Deposit date: | 2016-11-22 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Viral Immunoevasin Controls Innate Immunity by Targeting the Prototypical Natural Killer Cell Receptor Family.
Cell, 169, 2017
|
|
5YM8
 
 | |
5YM6
 
 | |
5WVI
 
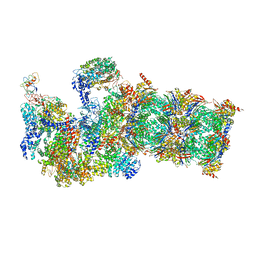 | | The resting state of yeast proteasome | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Ding, Z, Cong, Y. | | Deposit date: | 2016-12-25 | | Release date: | 2017-03-22 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | High-resolution cryo-EM structure of the proteasome in complex with ADP-AlFx
Cell Res., 27, 2017
|
|
5WVK
 
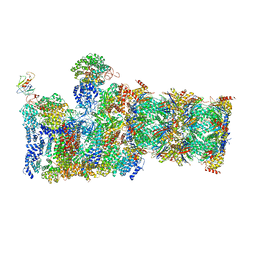 | | Yeast proteasome-ADP-AlFx | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Ding, Z, Cong, Y. | | Deposit date: | 2016-12-25 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | High-resolution cryo-EM structure of the proteasome in complex with ADP-AlFx
Cell Res., 27, 2017
|
|
8IXE
 
 | | GMPCPP-Alpha1C/Beta2A-microtubule decorated with kinesin seam region | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-1 heavy chain, ... | | Authors: | Zheng, W, Zhao, Q.Y, Diao, L, Bao, L, Cong, Y. | | Deposit date: | 2023-03-31 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM of alpha-tubulin isotype-containing microtubules revealed a contracted structure of alpha 4A/ beta 2A microtubules.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
8IXB
 
 | | GMPCPP-Alpha1A/Beta2A-microtubule decorated with kinesin seam region | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-1 heavy chain, ... | | Authors: | Zheng, W, Zhao, Q.Y, Diao, L, Bao, L, Cong, Y. | | Deposit date: | 2023-03-31 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM of alpha-tubulin isotype-containing microtubules revealed a contracted structure of alpha 4A/ beta 2A microtubules.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
8IXG
 
 | | GMPCPP-Alpha4A/Beta2A-microtubule decorated with kinesin seam region | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-1 heavy chain, ... | | Authors: | Zheng, W, Zhao, Q.Y, Diao, L, Bao, L, Cong, Y. | | Deposit date: | 2023-03-31 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM of alpha-tubulin isotype-containing microtubules revealed a contracted structure of alpha 4A/ beta 2A microtubules.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
8IXA
 
 | | GMPCPP-Alpha1A/Beta2A-microtubule decorated with kinesin non-seam region | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-1 heavy chain, ... | | Authors: | Zheng, W, Zhao, Q.Y, Diao, L, Bao, L, Cong, Y. | | Deposit date: | 2023-03-31 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM of alpha-tubulin isotype-containing microtubules revealed a contracted structure of alpha 4A/ beta 2A microtubules.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
8IXD
 
 | | GMPCPP-Alpha1C/Beta2A-microtubule decorated with kinesin non-seam region | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-1 heavy chain, ... | | Authors: | Zheng, W, Zhao, Q.Y, Diao, L, Bao, L, Cong, Y. | | Deposit date: | 2023-03-31 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM of alpha-tubulin isotype-containing microtubules revealed a contracted structure of alpha 4A/ beta 2A microtubules.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
8IXF
 
 | | GMPCPP-Alpha4A/Beta2A-microtubule decorated with kinesin non-seam region | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-1 heavy chain, ... | | Authors: | Zheng, W, Zhao, Q.Y, Diao, L, Bao, L, Cong, Y. | | Deposit date: | 2023-03-31 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM of alpha-tubulin isotype-containing microtubules revealed a contracted structure of alpha 4A/ beta 2A microtubules.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
5HIZ
 
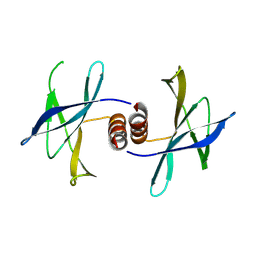 | | The structure of PEDV NSP9 | | Descriptor: | Non-structural protein 9 | | Authors: | Deng, F, Peng, G. | | Deposit date: | 2016-01-12 | | Release date: | 2017-01-25 | | Last modified: | 2019-07-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dimerization of Coronavirus nsp9 with Diverse Modes Enhances Its Nucleic Acid Binding Affinity.
J.Virol., 92, 2018
|
|
5HIY
 
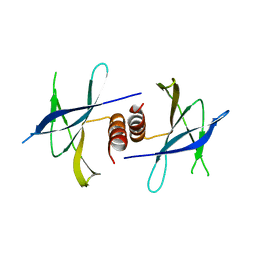 | | Crystal structure of PEDV NSP9 Mutant-C59A | | Descriptor: | Non-structural protein 9 | | Authors: | Deng, F, Peng, G. | | Deposit date: | 2016-01-12 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Dimerization of Coronavirus nsp9 with Diverse Modes Enhances Its Nucleic Acid Binding Affinity.
J.Virol., 92, 2018
|
|
8CDT
 
 | |
8CDS
 
 | | Crystal structure of the xhNup93-Nb4i VHH antibody | | Descriptor: | 1,2-ETHANEDIOL, 5-Nup93 inhibitory VHH antibody, DI(HYDROXYETHYL)ETHER | | Authors: | Guttler, T, Colom, M.S, Gorlich, D. | | Deposit date: | 2023-02-01 | | Release date: | 2024-02-21 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | A checkpoint function for Nup98 in nuclear pore formation suggested by novel inhibitory nanobodies.
Embo J., 43, 2024
|
|
7JPU
 
 | |
