6LEL
 
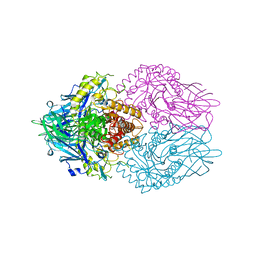 | | Structure of E. coli beta-glucuronidase complex with C6-hexyl uronic isofagomine | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-2-hexyl-4,5-bis(oxidanyl)piperidine-3-carboxylic acid, Beta-D-glucuronidase | | Authors: | Lin, H.-Y, Kuo, Y.-H, Lin, C.-H. | | Deposit date: | 2019-11-25 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Entropy-driven binding of gut bacterial beta-glucuronidase inhibitors ameliorates irinotecan-induced toxicity.
Commun Biol, 4, 2021
|
|
2KIU
 
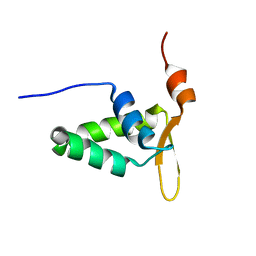 | |
9GIJ
 
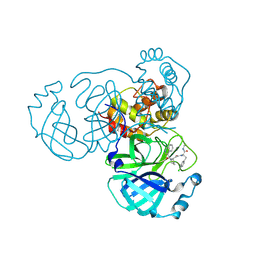 | | Crystal structure of SARS-CoV-2 Mpro with compound 5 | | Descriptor: | (2~{R})-3-(4-chlorophenyl)-2-[2-[(2~{R})-1-isoquinolin-4-ylcarbonylpyrrolidin-2-yl]ethanoyl-methyl-amino]-~{N}-methyl-propanamide, 3C-like proteinase nsp5 | | Authors: | Prasad, A, Schmitt, A, Preuss, F, Maskos, K, Wang, X, Gotchev, D, Konz Makino, D.L. | | Deposit date: | 2024-08-19 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.476 Å) | | Cite: | Rational Design of Macrocyclic Noncovalent Inhibitors of SARS-CoV-2 M pro from a DNA-Encoded Chemical Library Screening Hit That Demonstrate Potent Inhibition against Pan-Coronavirus Homologues and Nirmatrelvir-Resistant Variants.
J.Med.Chem., 2024
|
|
9GIL
 
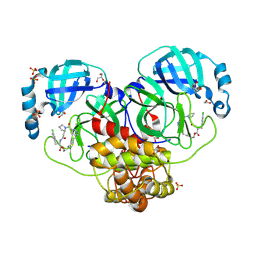 | | Crystal structure of SARS-CoV-2 Mpro with compound 12 | | Descriptor: | (7~{R},11~{R},19~{E})-11-[(4-chlorophenyl)methyl]-13-oxa-3,10,23-triazatricyclo[19.3.1.0^{3,7}]pentacosa-1(24),19,21(25),22-tetraene-2,9,12-trione, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Schmitt, A, Preuss, F, Prasad, A, Maskos, K, Wang, X, Gotchev, D, Konz Makino, D.L. | | Deposit date: | 2024-08-19 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Rational Design of Macrocyclic Noncovalent Inhibitors of SARS-CoV-2 M pro from a DNA-Encoded Chemical Library Screening Hit That Demonstrate Potent Inhibition against Pan-Coronavirus Homologues and Nirmatrelvir-Resistant Variants.
J.Med.Chem., 2024
|
|
6JZ6
 
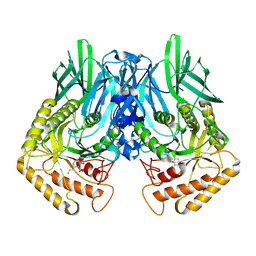 | | b-glucuronidase from Ruminococcus gnavus in complex with C6-substituted uronic isofagomine | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-4,5-bis(oxidanyl)-2-propyl-piperidine-3-carboxylic acid, Beta-glucuronidase | | Authors: | Dashnyam, P, Lin, H.Y. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.605 Å) | | Cite: | Substituent Position of Iminocyclitols Determines the Potency and Selectivity for Gut Microbial Xenobiotic-Reactivating Enzymes.
J.Med.Chem., 63, 2020
|
|
6L6E
 
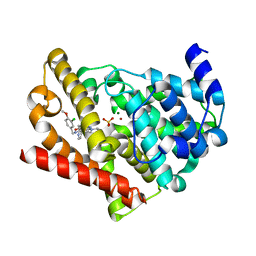 | | Human PDE5 catalytic core in complex with avanafil | | Descriptor: | 4-[(3-chloranyl-4-methoxy-phenyl)methylamino]-2-[(2S)-2-(hydroxymethyl)pyrrolidin-1-yl]-N-(pyrimidin-2-ylmethyl)pyrimid ine-5-carboxamide, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Hsieh, C.M, Chan, N.L. | | Deposit date: | 2019-10-28 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of Human Phosphodiesterase 5A1 Complexed with Avanafil Reveals Molecular Basis of Isoform Selectivity and Guidelines for Targeting alpha-Helix Backbone Oxygen by Halogen Bonding.
J.Med.Chem., 63, 2020
|
|
1RLA
 
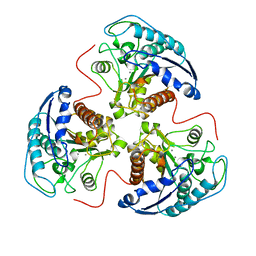 | | THREE-DIMENSIONAL STRUCTURE OF RAT LIVER ARGINASE, THE BINUCLEAR MANGANESE METALLOENZYME OF THE UREA CYCLE | | Descriptor: | ARGINASE, MANGANESE (II) ION | | Authors: | Kanyo, Z, Scolnick, L, Ash, D, Christianson, D.W. | | Deposit date: | 1996-08-15 | | Release date: | 1997-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a unique binuclear manganese cluster in arginase.
Nature, 383, 1996
|
|
3GRL
 
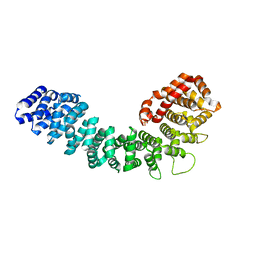 | |
3GQ2
 
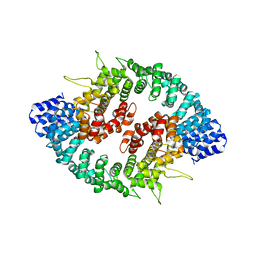 | |
6LDC
 
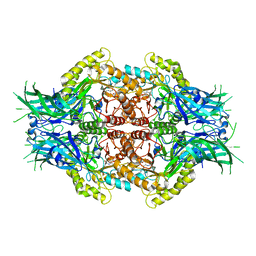 | | Structure of Bifidobacterium dentium beta-glucuronidase complexed with C6-nonyl uronic isofagomine | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-2-nonyl-4,5-bis(oxidanyl)piperidine-3-carboxylic acid, LacZ1 Beta-galactosidase | | Authors: | Lin, H.-Y, Hsieh, T.-J, Lin, C.-H. | | Deposit date: | 2019-11-20 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.181 Å) | | Cite: | Entropy-driven binding of gut bacterial beta-glucuronidase inhibitors ameliorates irinotecan-induced toxicity.
Commun Biol, 4, 2021
|
|
6J03
 
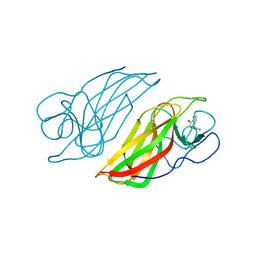 | | Crystal structure of a cyclase mutant in apo form | | Descriptor: | 12-epi-hapalindole U synthase, CALCIUM ION, amino({3-[(3S,8aS)-1,4-dioxooctahydropyrrolo[1,2-a]pyrazin-3-yl]propyl}amino)methaniminium | | Authors: | Tang, X.K, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural insights into the calcium dependence of Stig cyclases.
Rsc Adv, 9, 2019
|
|
2HH3
 
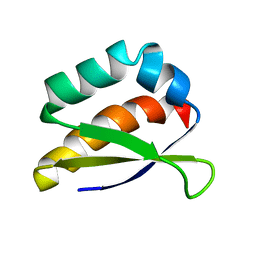 | | Solution structure of the third KH domain of KSRP | | Descriptor: | KH-type splicing regulatory protein | | Authors: | Garcia-Mayoral, M.F. | | Deposit date: | 2006-06-27 | | Release date: | 2007-05-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Structure of the C-Terminal KH Domains of KSRP Reveals a Noncanonical Motif Important for mRNA Degradation.
Structure, 15, 2007
|
|
2HH2
 
 | |
5J7J
 
 | |
6JZ8
 
 | | b-glucuronidase from Ruminococcus gnavus in complex with D-glucaro 1,5-lactone | | Descriptor: | (2S,3S,4S,5R)-3,4,5-trihydroxy-6-oxo-oxane-2-carboxylic acid, Beta-glucuronidase | | Authors: | Dashnyam, P, Lin, H.Y. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.583 Å) | | Cite: | Substituent Position of Iminocyclitols Determines the Potency and Selectivity for Gut Microbial Xenobiotic-Reactivating Enzymes.
J.Med.Chem., 63, 2020
|
|
6JZ5
 
 | |
6JZ3
 
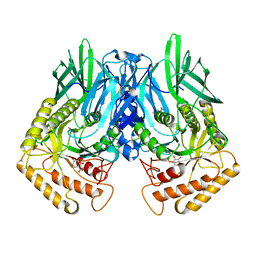 | | b-glucuronidase from Ruminococcus gnavus in complex with uronic deoxynojirimycin | | Descriptor: | (2~{S},3~{R},4~{R},5~{S})-3,4,5-tris(oxidanyl)piperidine-2-carboxylic acid, (4R)-2-METHYLPENTANE-2,4-DIOL, Beta-glucuronidase | | Authors: | Dashnyam, P, Lin, H.Y. | | Deposit date: | 2019-04-30 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Substituent Position of Iminocyclitols Determines the Potency and Selectivity for Gut Microbial Xenobiotic-Reactivating Enzymes.
J.Med.Chem., 63, 2020
|
|
6JZ2
 
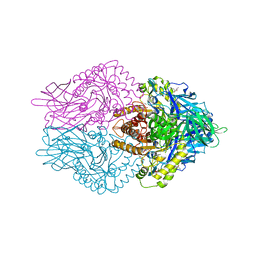 | | b-glucuronidase from Ruminococcus gnavus in complex with uronic isofagomine at 1.3 Angstroms resolution | | Descriptor: | (3S,4R,5R)-4,5-dihydroxypiperidine-3-carboxylic acid, (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | Authors: | Dashnyam, P, Lin, H.Y. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Substituent Position of Iminocyclitols Determines the Potency and Selectivity for Gut Microbial Xenobiotic-Reactivating Enzymes.
J.Med.Chem., 63, 2020
|
|
6JZ4
 
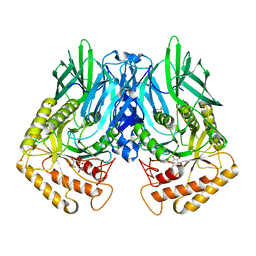 | | b-glucuronidase from Ruminococcus gnavus in complex with D-glucaro-d-lactam | | Descriptor: | (2S,3R,4S,5R)-3,4,5-trihydroxy-6-oxopiperidine-2-carboxylic acid, (4R)-2-METHYLPENTANE-2,4-DIOL, Beta-glucuronidase | | Authors: | Dashnyam, P, Lin, H.Y. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | Substituent Position of Iminocyclitols Determines the Potency and Selectivity for Gut Microbial Xenobiotic-Reactivating Enzymes.
J.Med.Chem., 63, 2020
|
|
6JZ1
 
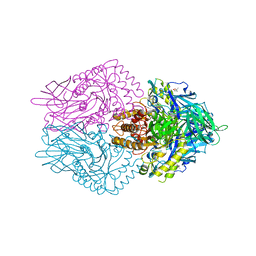 | |
6JZ7
 
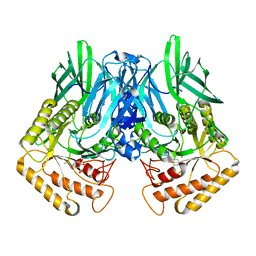 | |
5EK3
 
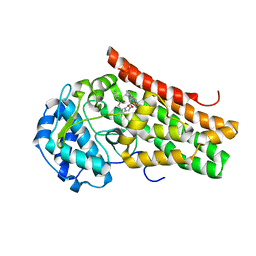 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with NLG919 analogue | | Descriptor: | (1~{R})-1-cyclohexyl-2-[(5~{S})-5~{H}-imidazo[1,5-b]isoindol-5-yl]ethanol, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Peng, Y.H, Wu, J.S, Wu, S.Y. | | Deposit date: | 2015-11-03 | | Release date: | 2015-12-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | Important Hydrogen Bond Networks in Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitor Design Revealed by Crystal Structures of Imidazoleisoindole Derivatives with IDO1
J.Med.Chem., 59, 2016
|
|
5EK2
 
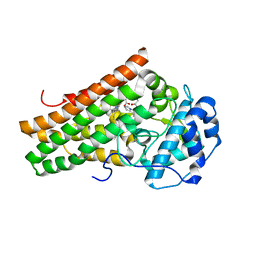 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with NLG919 analogue | | Descriptor: | 1-cyclohexyl-2-[(5~{S})-6-fluoranyl-5~{H}-imidazo[1,5-b]isoindol-5-yl]ethanone, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Peng, Y.H, Wu, J.S, Wu, S.Y. | | Deposit date: | 2015-11-03 | | Release date: | 2015-12-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Important Hydrogen Bond Networks in Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitor Design Revealed by Crystal Structures of Imidazoleisoindole Derivatives with IDO1
J.Med.Chem., 59, 2016
|
|
5ETW
 
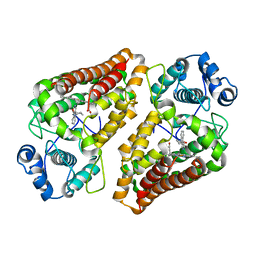 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with NLG919 analogue | | Descriptor: | (1~{R})-1-cyclohexyl-2-pyrido[3,4-b]indol-9-yl-ethanol, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, S.Y, Peng, Y.H, Wu, J.S. | | Deposit date: | 2015-11-18 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Important Hydrogen Bond Networks in Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitor Design Revealed by Crystal Structures of Imidazoleisoindole Derivatives with IDO1.
J.Med.Chem., 59, 2016
|
|
5EK4
 
 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with NLG919 analogue | | Descriptor: | (1~{R})-1-cyclohexyl-2-[(5~{S})-6-fluoranyl-5~{H}-imidazo[1,5-b]isoindol-5-yl]ethanol, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, S.Y, Peng, Y.H, Wu, J.S. | | Deposit date: | 2015-11-03 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Important Hydrogen Bond Networks in Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitor Design Revealed by Crystal Structures of Imidazoleisoindole Derivatives with IDO1
J.Med.Chem., 59, 2016
|
|
