7ELJ
 
 | |
6JXD
 
 | |
8GJV
 
 | |
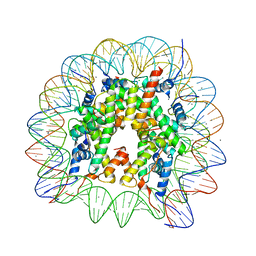
6K1I
 
 | | Human nucleosome core particle with gammaH2A.X variant | | Descriptor: | CHLORIDE ION, DNA (147-MER), Histone H2AX, ... | | Authors: | Sharma, D, De Falco, L, Davey, C.A. | | Deposit date: | 2019-05-10 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | PARP1 exhibits enhanced association and catalytic efficiency with gamma H2A.X-nucleosome.
Nat Commun, 10, 2019
|
|
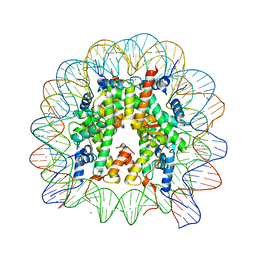
6K1K
 
 | | Human nucleosome core particle with H2A.X S139E variant | | Descriptor: | CHLORIDE ION, DNA (145-MER), Histone H2AX, ... | | Authors: | Sharma, D, De Falco, L, Davey, C.A. | | Deposit date: | 2019-05-10 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | PARP1 exhibits enhanced association and catalytic efficiency with gamma H2A.X-nucleosome.
Nat Commun, 10, 2019
|
|
6K1J
 
 | | Human nucleosome core particle with H2A.X variant | | Descriptor: | CHLORIDE ION, DNA (145-MER), Histone H2AX, ... | | Authors: | Sharma, D, De Falco, L, Davey, C.A. | | Deposit date: | 2019-05-10 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | PARP1 exhibits enhanced association and catalytic efficiency with gamma H2A.X-nucleosome.
Nat Commun, 10, 2019
|
|
6MTO
 
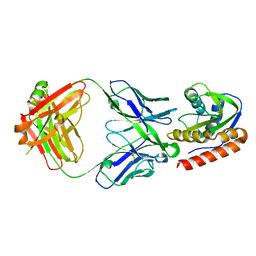 | | Crystal structure of VRC42.01 Fab in complex with T117-F MPER scaffold | | Descriptor: | Antibody VRC42.01 Fab heavy chain, Antibody VRC42.01 Fab light chain, VRC42 epitope T117-F scaffold | | Authors: | Kwon, Y.D, Druz, A, Law, W.H, Peng, D, Zhang, B, Doria-Rose, N.A, Kwong, P.D. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.634 Å) | | Cite: | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
6MTR
 
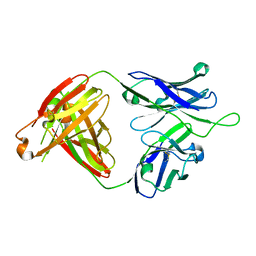 | | Crystal structure of VRC43.01 Fab | | Descriptor: | Antibody VRC43.01 Fab heavy chain, Antibody VRC43.01 Fab light chain | | Authors: | Kwon, Y.D, Druz, A, Law, W.H, Peng, D, Zhang, B, Doria-Rose, N.A, Kwong, P.D. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
6MTS
 
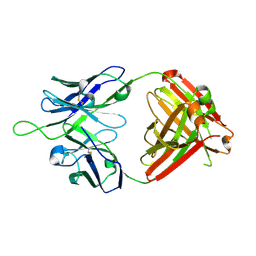 | | Crystal structure of VRC43.03 Fab | | Descriptor: | Antibody VRC43.03 Fab heavy chain, Antibody VRC43.03 Fab light chain | | Authors: | Kwon, Y.D, Druz, A, Law, W.H, Peng, D, Doria-Rose, N.A, Kwong, P.D. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.437 Å) | | Cite: | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
6MTP
 
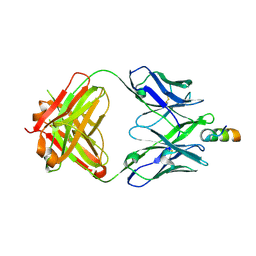 | | Crystal structure of VRC42.04 Fab in complex with gp41 peptide | | Descriptor: | Antibody VRC42.04 Fab heavy chain, Antibody VRC42.04 Fab light chain, RV217 founder virus gp41 peptide | | Authors: | Kwon, Y.D, Druz, A, Law, W.H, Peng, D, Zhang, B, Doria-Rose, N.A, Kwong, P.D. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.036 Å) | | Cite: | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
6MTT
 
 | | Crystal structure of VRC46.01 Fab in complex with gp41 peptide | | Descriptor: | Antibody VRC46.01 Fab heavy chain, Antibody VRC46.01 Fb light chain, RV217 founder virus gp41 peptide | | Authors: | Kwon, Y.D, Druz, A, Law, W.H, Peng, D, Veradi, R, Doria-Rose, N.A, Kwong, P.D. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
5U3P
 
 | | Crystal Structure of DH511.4 Fab | | Descriptor: | DH511.4 Fab Heavy Chain, DH511.4 Fab Light Chain | | Authors: | Nicely, N.I, Williams, L.D, Ofek, G, Haynes, B.F. | | Deposit date: | 2016-12-02 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Potent and broad HIV-neutralizing antibodies in memory B cells and plasma.
Sci Immunol, 2, 2017
|
|
5U3J
 
 | | Crystal Structure of DH511.1 Fab in Complex with HIV-1 gp41 MPER Peptide | | Descriptor: | DH511.1 Heavy Chain, DH511.1 Light Chain, gp41 MPER peptide | | Authors: | Ofek, G, Wu, L, Lougheed, C.S, Williams, L.D, Nicely, N.I, Haynes, B.F. | | Deposit date: | 2016-12-02 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Potent and broad HIV-neutralizing antibodies in memory B cells and plasma.
Sci Immunol, 2, 2017
|
|
5U3K
 
 | | Crystal Structure of DH511.2 Fab in Complex with HIV-1 gp41 MPER 662-683 Peptide | | Descriptor: | CALCIUM ION, CHLORIDE ION, DH511.2 Fab Heavy Chain, ... | | Authors: | Ofek, G, Wu, L, Lougheed, C.S, Williams, L.D, Nicely, N.I, Haynes, B.F. | | Deposit date: | 2016-12-02 | | Release date: | 2017-02-15 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.637 Å) | | Cite: | Potent and broad HIV-neutralizing antibodies in memory B cells and plasma.
Sci Immunol, 2, 2017
|
|
5U3O
 
 | | Crystal Structure of DH511.2_K3 Fab in Complex with HIV-1 gp41 MPER Peptide | | Descriptor: | DH511.2_K3 Fab Heavy Chain, DH511.2_K3 Fab Light Chain, gp41 MPER peptide | | Authors: | Ofek, G, Wu, L, Lougheed, C.S, Williams, L.D, Nicely, N.I, Haynes, B.F. | | Deposit date: | 2016-12-02 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | Potent and broad HIV-neutralizing antibodies in memory B cells and plasma.
Sci Immunol, 2, 2017
|
|
5U3N
 
 | | Crystal Structure of DH511.12P Fab in Complex with HIV-1 gp41 MPER Peptide | | Descriptor: | DH511.12P Fab Heavy Chain, DH511.12P Fab Light Chain, gp41 MPER peptide | | Authors: | Ofek, G, Wu, L, Lougheed, C.S, Williams, L.D, Nicely, N.I, Haynes, B.F. | | Deposit date: | 2016-12-02 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent and broad HIV-neutralizing antibodies in memory B cells and plasma.
Sci Immunol, 2, 2017
|
|
5U3L
 
 | | Crystal Structure of DH511.2 Fab in Complex with HIV-1 gp41 MPER 670-683 Peptide | | Descriptor: | DH511.2 Fab Heavy Chain, DH511.2 Fab Light Chain, gp41 MPER peptide | | Authors: | Ofek, G, Wu, L, Lougheed, C.S, Williams, L.D, Nicely, N.I, Haynes, B.F. | | Deposit date: | 2016-12-02 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.165 Å) | | Cite: | Potent and broad HIV-neutralizing antibodies in memory B cells and plasma.
Sci Immunol, 2, 2017
|
|
5U3M
 
 | | Crystal Structure of DH511.11P Fab in Complex with HIV-1 gp41 MPER Peptide | | Descriptor: | DH511.11P Fab Heavy Chain, DH511.11P Fab Light Chain, gp41 MPER peptide | | Authors: | Ofek, G, Wu, L, Lougheed, C.S, Williams, L.D, Nicely, N.I, Haynes, B.F. | | Deposit date: | 2016-12-02 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.418 Å) | | Cite: | Potent and broad HIV-neutralizing antibodies in memory B cells and plasma.
Sci Immunol, 2, 2017
|
|
6O3O
 
 | | Structure of human DNAM-1 (CD226) bound to nectin-like protein-5 (necl-5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD226 antigen, ... | | Authors: | Deuss, F.A, Watson, G.M, Rossjohn, J, Berry, R. | | Deposit date: | 2019-02-27 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the recognition of nectin-like protein-5 by the human-activating immune receptor, DNAM-1.
J.Biol.Chem., 294, 2019
|
|
5A2H
 
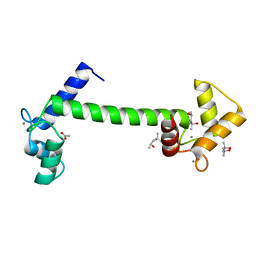 | | Crystal Structure of Arabidopsis thaliana Calmodulin-7 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Kumar, S, Gourinath, S. | | Deposit date: | 2015-05-19 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure of Arabidopsis Thaliana Calmodulin7 and Insight Into its Mode of DNA Binding.
FEBS Lett., 590, 2016
|
|
6CJJ
 
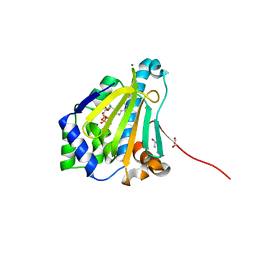 | | Candida albicans Hsp90 nucleotide binding domain in complex with ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Heat shock protein 90 homolog, ... | | Authors: | Hutchinson, A, Loppnau, P, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Hui, R, STRUCTURAL GENOMICS CONSORTIUM, S.G.C, Pizarro, J.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-26 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis for species-selective targeting of Hsp90 in a pathogenic fungus.
Nat Commun, 10, 2019
|
|
6CJS
 
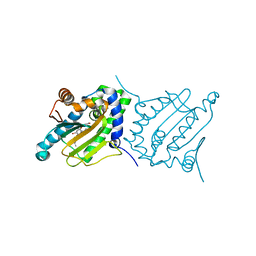 | |
6CJP
 
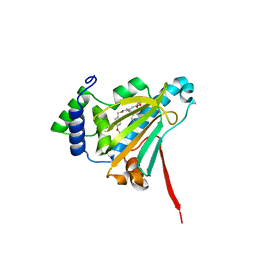 | | Candida albicans Hsp90 nucleotide binding domain in complex with radicicol | | Descriptor: | (1aR,2Z,4E,6E,14R,15aR)-9,11-dihydroxy-6-{[(4-methoxyphenyl)methoxy]imino}-14-methyl-1a,6,7,14,15,15a-hexahydro-12H-oxireno[e][2]benzoxacyclotetradecin-12-one, Heat shock protein 90 homolog | | Authors: | Nation, C, Pizarro, J.C. | | Deposit date: | 2018-02-26 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for species-selective targeting of Hsp90 in a pathogenic fungus.
Nat Commun, 10, 2019
|
|
6CJL
 
 | |
7BX2
 
 | |
