1N32
 
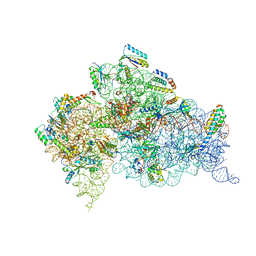 | | Structure of the Thermus thermophilus 30S ribosomal subunit bound to codon and near-cognate transfer RNA anticodon stem-loop mismatched at the first codon position at the a site with paromomycin | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Ogle, J.M, Murphy IV, F.V, Tarry, M.J, Ramakrishnan, V. | | Deposit date: | 2002-10-25 | | Release date: | 2002-11-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Selection of tRNA by the Ribosome Requires a Transition from an Open to a Closed Form
Cell(Cambridge,Mass.), 111, 2002
|
|
1N36
 
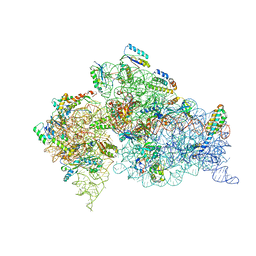 | | Structure of the Thermus thermophilus 30S ribosomal subunit in the presence of crystallographically disordered codon and near-cognate transfer RNA anticodon stem-loop mismatched at the second codon position | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Ogle, J.M, Murphy IV, F.V, Tarry, M.J, Ramakrishnan, V. | | Deposit date: | 2002-10-25 | | Release date: | 2002-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Selection of tRNA by the Ribosome Requires a Transition from an Open to a Closed Form
Cell(Cambridge,Mass.), 111, 2002
|
|
6ZOZ
 
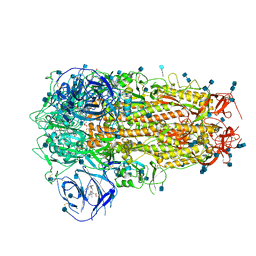 | | Structure of Disulphide-stabilized SARS-CoV-2 Spike Protein Trimer (x1 disulphide-bond mutant, S383C, D985C, K986P, V987P, single Arg S1/S2 cleavage site) in Locked State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7YE9
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
6ZP1
 
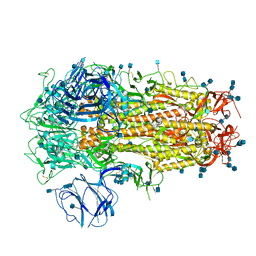 | | Structure of SARS-CoV-2 Spike Protein Trimer (K986P, V987P, single Arg S1/S2 cleavage site) in Closed State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZOY
 
 | | Structure of Disulphide-stabilized SARS-CoV-2 Spike Protein Trimer (x1 disulphide-bond mutant, S383C, D985C, K986P, V987P, single Arg S1/S2 cleavage site) in Closed State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZP0
 
 | | Structure of SARS-CoV-2 Spike Protein Trimer (single Arg S1/S2 cleavage site) in Closed State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZOX
 
 | | Structure of Disulphide-stabilized SARS-CoV-2 Spike Protein Trimer (x2 disulphide-bond mutant, G413C, V987C, single Arg S1/S2 cleavage site) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZP2
 
 | | Structure of SARS-CoV-2 Spike Protein Trimer (K986P, V987P, single Arg S1/S2 cleavage site) in Locked State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7YDY
 
 | | SARS-CoV-2 Spike (6P) in complex with 1 R1-32 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-04 | | Release date: | 2022-08-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.75 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YEG
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YDI
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2, focused refinement of RBD region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32, Light chain of R1-32, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YE5
 
 | | SARS-CoV-2 Spike (6P) in complex with 2 R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.75 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
8ARF
 
 | |
1T1M
 
 | | Binding position of ribosome recycling factor (RRF) on the E. coli 70S ribosome | | Descriptor: | 42-mer fragment of double helix from 16S rRNA, dodecamer fragment of double helix from 23S rRNA, ribosome recycling factor | | Authors: | Agrawal, R.K, Sharma, M.R, Kiel, M.C, Hirokawa, G, Booth, T.M, Spahn, C.M, Grassucci, R.A, Kaji, A, Frank, J. | | Deposit date: | 2004-04-16 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: Functional implications
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1T1O
 
 | | Components of the control 70S ribosome to provide reference for the RRF binding site | | Descriptor: | 19-mer fragment of the 23S rRNA, 42-mer fragment of double helix from 16S rRNA, dodecamer fragment of double helix from 23S rRNA | | Authors: | Agrawal, R.K, Sharma, M.R, Kiel, M.C, Hirokawa, G, Booth, T.M, Spahn, C.M, Grassucci, R.A, Kaji, A, Frank, J. | | Deposit date: | 2004-04-16 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: Functional implications
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1X18
 
 | | Contact sites of ERA GTPase on the THERMUS THERMOPHILUS 30S SUBUNIT | | Descriptor: | 30S ribosomal protein S11, 30S ribosomal protein S18, 30S ribosomal protein S2, ... | | Authors: | Sharma, M.R, Barat, C, Agrawal, R.K. | | Deposit date: | 2005-04-02 | | Release date: | 2005-05-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Interaction of Era with the 30S Ribosomal Subunit Implications for 30S Subunit Assembly
Mol.Cell, 18, 2005
|
|
1X1L
 
 | | Interaction of ERA,a GTPase protein, with the 3'minor domain of the 16S rRNA within the THERMUS THERMOPHILUS 30S subunit. | | Descriptor: | GTP-binding protein era, RNA (130-MER) | | Authors: | Sharma, M.R, Barat, C, Agrawal, R.K. | | Deposit date: | 2005-04-06 | | Release date: | 2005-05-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Interaction of Era with the 30S Ribosomal Subunit Implications for 30S Subunit Assembly
Mol.Cell, 18, 2005
|
|
2R1G
 
 | | Coordinates of the thermus thermophilus 30S components neighboring RbfA as obtained by fitting into the CRYO-EM map of A 30S-RBFA complex | | Descriptor: | 16S RIBOSOMAL RNA HELIX 1, 16S RIBOSOMAL RNA HELIX 18, 16S RIBOSOMAL RNA HELIX 27, ... | | Authors: | Datta, P.P, Wilson, D.N, Kawazoe, M, Swami, N.K, Kaminishi, T, Sharma, M.R, Booth, T.M, Takemoto, C, Fucini, P, Yokoyama, S, Agrawal, R.K. | | Deposit date: | 2007-08-22 | | Release date: | 2008-03-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (12.5 Å) | | Cite: | Structural aspects of RbfA action during small ribosomal subunit assembly.
Mol.Cell, 28, 2007
|
|
