7SRV
 
 | | Metal dependent activation of Plasmodium falciparum M17 aminopeptidase (inactive form), spacegroup P22121 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Webb, C.T, McGowan, S. | | Deposit date: | 2021-11-08 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A metal ion-dependent conformational switch modulates activity of the Plasmodium M17 aminopeptidase.
J.Biol.Chem., 298, 2022
|
|
3TUU
 
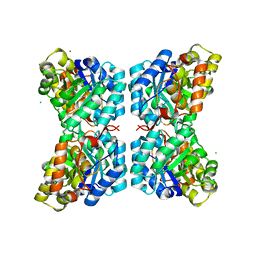 | | Structure of dihydrodipicolinate synthase from the common grapevine | | Descriptor: | BROMIDE ION, CHLORIDE ION, dihydrodipicolinate synthase | | Authors: | Perugini, M.A, Dobson, R.C, Atkinson, S.C. | | Deposit date: | 2011-09-19 | | Release date: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal, Solution and In silico Structural Studies of Dihydrodipicolinate Synthase from the Common Grapevine.
Plos One, 7, 2012
|
|
6P4B
 
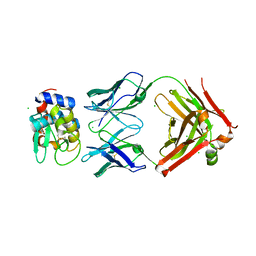 | | HyHEL10 fab variant HyHEL10-4x (heavy chain mutations L4F, Y33H, S56N, and Y58F) bound to hen egg lysozyme variant HEL2x-flex (mutations R21Q, R73E, C76S, and C94S) | | Descriptor: | CHLORIDE ION, HyHEL10 Fab heavy chain, HyHEL10 Fab light chain, ... | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2019-05-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational diversity facilitates antibody mutation trajectories and discrimination between foreign and self-antigens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P4C
 
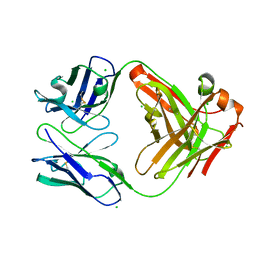 | | HyHEL10 Fab carrying four heavy chain mutations (HyHEL10-4x): L4F, Y33H, S56N, and Y58F | | Descriptor: | CHLORIDE ION, HyHEL10 Fab heavy chain, HyHEL10 Fab light chain | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2019-05-27 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational diversity facilitates antibody mutation trajectories and discrimination between foreign and self-antigens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P4D
 
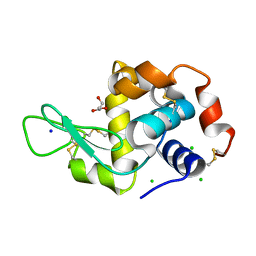 | | Hen egg lysozyme (HEL) containing three point mutations (HEL3x): R21Q, R73E, and D101R | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2019-05-27 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Conformational diversity facilitates antibody mutation trajectories and discrimination between foreign and self-antigens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P4A
 
 | |
1XU8
 
 | | The 2.8 A structure of a tumour suppressing serpin | | Descriptor: | Maspin, SULFATE ION | | Authors: | Irving, J.A, Law, R.H, Ruzyla, K, Bashtannyk-Puhalovich, T.A, Kim, N, Worrall, D.M, Rossjohn, J, Whisstock, J.C. | | Deposit date: | 2004-10-25 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The high resolution crystal structure of the human tumor suppressor maspin reveals a novel conformational switch in the G-helix.
J.Biol.Chem., 280, 2005
|
|
5V3B
 
 | |
5V3P
 
 | |
3KQZ
 
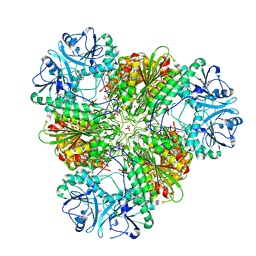 | | Structure of a protease 2 | | Descriptor: | CARBONATE ION, M17 leucyl aminopeptidase, NONAETHYLENE GLYCOL, ... | | Authors: | McGowan, S, Whisstock, J.C. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of the Plasmodium falciparum M17 aminopeptidase and significance for the design of drugs targeting the neutral exopeptidases
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KQX
 
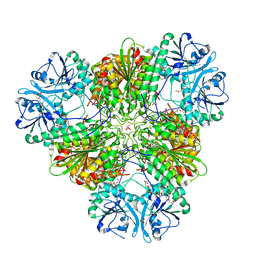 | | Structure of a protease 1 | | Descriptor: | CARBONATE ION, M17 leucyl aminopeptidase, NONAETHYLENE GLYCOL, ... | | Authors: | McGowan, S, Whisstock, J.C. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of the Plasmodium falciparum M17 aminopeptidase and significance for the design of drugs targeting the neutral exopeptidases
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KR5
 
 | | Structure of a protease 4 | | Descriptor: | (2S)-3-[(R)-[(1S)-1-amino-3-phenylpropyl](hydroxy)phosphoryl]-2-benzylpropanoic acid, CARBONATE ION, M17 leucyl aminopeptidase, ... | | Authors: | McGowan, S, Whisstock, J.C. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure of the Plasmodium falciparum M17 aminopeptidase and significance for the design of drugs targeting the neutral exopeptidases
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KR4
 
 | | Structure of a protease 3 | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, CARBONATE ION, M17 leucyl aminopeptidase, ... | | Authors: | McGowan, S, Whisstock, J.C. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Plasmodium falciparum M17 aminopeptidase and significance for the design of drugs targeting the neutral exopeptidases
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4UZM
 
 | |
4XAY
 
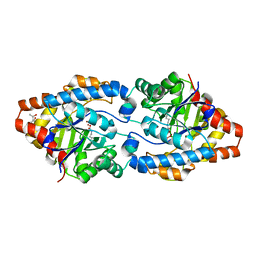 | | Cycles of destabilization and repair underlie evolutionary transitions in enzymes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-R8, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-16 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4XAZ
 
 | | Cycles of destabilization and repair underlie evolutionary transitions in enzymes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Phosphotriesterase variant PTE-R18, ZINC ION | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-16 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4XD4
 
 | | Phosphotriesterase variant E2b | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-R3, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-19 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4XAG
 
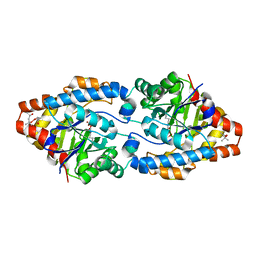 | | Cycles of destabilization and repair underlie the evolution of new enzyme function | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-R6, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-14 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4XD3
 
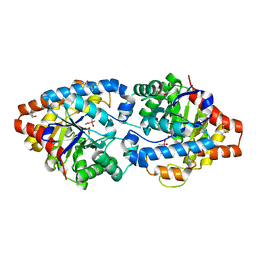 | | Phosphotriesterase variant E3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-E1, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-19 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4XD6
 
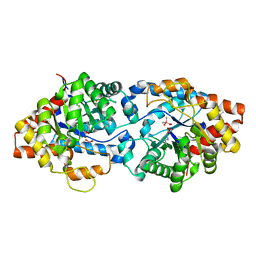 | | Phosphotriesterase Variant E2a | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-E2, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-19 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4XD5
 
 | | Phosphotriesterase variant R2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-R2, ... | | Authors: | Campbell, E, Kaltenbach, M, Tokuriki, N, Jackson, C.J. | | Deposit date: | 2014-12-19 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4XAF
 
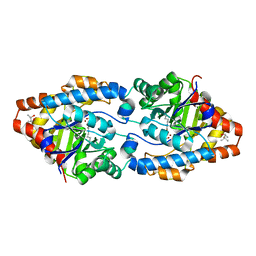 | | Cycles of destabilization and repair underlie evolutionary transitions in enzymes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-R1, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-14 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
7KWW
 
 | | X-ray Crystal Structure of PlyCB Mutant K59H | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PlyCB | | Authors: | Williams, D.E, Broendum, S.S, Hayes, B.K, Drinkwater, N, McGowan, S. | | Deposit date: | 2020-12-02 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High avidity drives the interaction between the streptococcal C1 phage endolysin, PlyC, with the cell surface carbohydrates of Group A Streptococcus.
Mol.Microbiol., 116, 2021
|
|
7KWT
 
 | | X-ray Crystal Structure of PlyCB Mutant Y28H | | Descriptor: | PlyCB | | Authors: | Williams, D.E, Broendum, S.S, Hayes, B.K, Drinkwater, N, McGowan, S. | | Deposit date: | 2020-12-02 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | High avidity drives the interaction between the streptococcal C1 phage endolysin, PlyC, with the cell surface carbohydrates of Group A Streptococcus.
Mol.Microbiol., 116, 2021
|
|
7KWY
 
 | | X-ray Crystal Structure of PlyCB Mutant R66K | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PlyCB | | Authors: | Williams, D.E, Broendum, S.S, Hayes, B.K, Drinkwater, N, McGowan, S. | | Deposit date: | 2020-12-02 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High avidity drives the interaction between the streptococcal C1 phage endolysin, PlyC, with the cell surface carbohydrates of Group A Streptococcus.
Mol.Microbiol., 116, 2021
|
|
