4Z03
 
 | | C. bescii Family 3 pectate lyase double mutant K108A in complex with trigalacturonic acid | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2015-03-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The catalytic mechanism and unique low pH optimum of Caldicellulosiruptor bescii family 3 pectate lyase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4YZX
 
 | | C. bescii Family 3 pectate lyase double mutant K108A/D107N in complex with trigalacturonic acid | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2015-03-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The catalytic mechanism and unique low pH optimum of Caldicellulosiruptor bescii family 3 pectate lyase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5BV9
 
 | |
4YZ0
 
 | | C. bescii Family 3 pectate lyase double mutant K108A/E39Q in complex with trigalacturonic acid | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2015-03-24 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The catalytic mechanism and unique low pH optimum of Caldicellulosiruptor bescii family 3 pectate lyase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4YZQ
 
 | | C. bescii Family 3 pectate lyase double mutant K108A/Q111N in complex with trigalacturonic acid | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Pectate lyase, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2015-03-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The catalytic mechanism and unique low pH optimum of Caldicellulosiruptor bescii family 3 pectate lyase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7ZEV
 
 | | Free form of extended Cyp33-RRM | | Descriptor: | Peptidyl-prolyl cis-trans isomerase E | | Authors: | Blatter, M, Allain, F, Meylan, C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | RNA binding induces an allosteric switch in Cyp33 to repress MLL1-mediated transcription.
Sci Adv, 9, 2023
|
|
7ZEW
 
 | | Complex Cyp33-RRM : AAUAAA RNA | | Descriptor: | Peptidyl-prolyl cis-trans isomerase E, RNA (5'-R(*AP*AP*UP*AP*AP*A)-3') | | Authors: | Blatter, M, Allain, F, Meylan, C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | RNA binding induces an allosteric switch in Cyp33 to repress MLL1-mediated transcription.
Sci Adv, 9, 2023
|
|
7ZEZ
 
 | | Trimolecular complex Cyp33-RRMdelta alpha : MLL1-PHD3 : H3K4me3 | | Descriptor: | Histone H3, Isoform 3 of Peptidyl-prolyl cis-trans isomerase E, MLL cleavage product N320, ... | | Authors: | Blatter, M, Allain, F, Meylan, C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2023-05-03 | | Method: | SOLUTION NMR | | Cite: | RNA binding induces an allosteric switch in Cyp33 to repress MLL1-mediated transcription.
Sci Adv, 9, 2023
|
|
7ZEX
 
 | | Complex Cyp33-RRMdelta alpha : UAAUGUCG RNA | | Descriptor: | Isoform 3 of Peptidyl-prolyl cis-trans isomerase E, RNA (5'-R(*UP*AP*AP*UP*GP*UP*CP*G)-3') | | Authors: | Blatter, M, Allain, F, Meylan, C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-05-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | RNA binding induces an allosteric switch in Cyp33 to repress MLL1-mediated transcription.
Sci Adv, 9, 2023
|
|
5TMP
 
 | |
7ZEY
 
 | | Complex Cyp33-RRM : MLL1-PHD3 | | Descriptor: | MLL cleavage product N320, Peptidyl-prolyl cis-trans isomerase E, ZINC ION | | Authors: | Blatter, M, Allain, F, Meylan, C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | RNA binding induces an allosteric switch in Cyp33 to repress MLL1-mediated transcription.
Sci Adv, 9, 2023
|
|
3M76
 
 | |
3M73
 
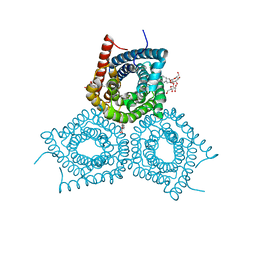 | |
4EW9
 
 | | The liganded structure of C. bescii family 3 pectate lyase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-deoxy-beta-L-threo-hex-4-enopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid, ACETATE ION, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2012-04-26 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure and mode of action of Caldicellulosiruptor bescii family 3 pectate lyase in biomass deconstruction.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
3M75
 
 | |
3PJZ
 
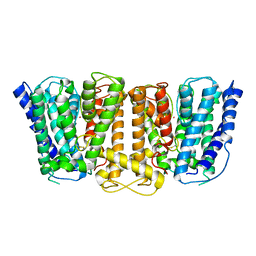 | | Crystal Structure of the Potassium Transporter TrkH from Vibrio parahaemolyticus | | Descriptor: | POTASSIUM ION, Potassium uptake protein TrkH | | Authors: | Cao, Y, Jin, X, Huang, H, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2010-11-10 | | Release date: | 2011-01-19 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.506 Å) | | Cite: | Crystal structure of a potassium ion transporter, TrkH.
Nature, 471, 2011
|
|
4PGR
 
 | |
4PGV
 
 | |
4PGS
 
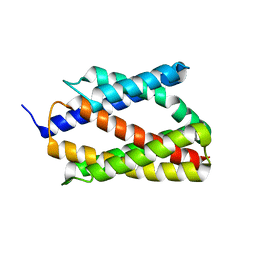 | |
4PGU
 
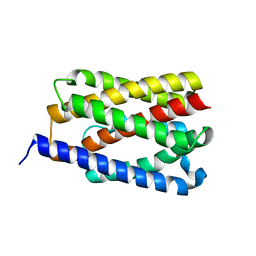 | |
4PGW
 
 | |
4RYQ
 
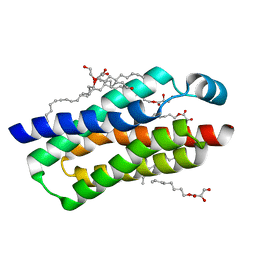 | | Crystal structure of BcTSPO, type 2 at 1.7 Angstrom | | Descriptor: | Integral membrane protein, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Guo, Y, Liu, Q, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-12-16 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protein structure. Structure and activity of tryptophan-rich TSPO proteins.
Science, 347, 2015
|
|
4RYI
 
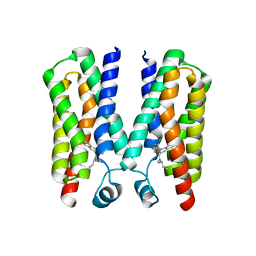 | |
4RYN
 
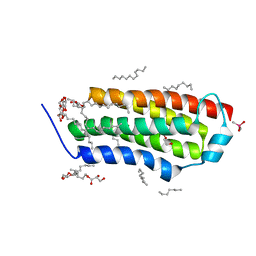 | | Crystal structure of BcTSPO, type1 monomer | | Descriptor: | CACODYLATE ION, DODECYL-ALPHA-D-MALTOSIDE, Integral membrane protein, ... | | Authors: | Guo, Y, Liu, Q, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-12-15 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Protein structure. Structure and activity of tryptophan-rich TSPO proteins.
Science, 347, 2015
|
|
4RYJ
 
 | |
