7UPX
 
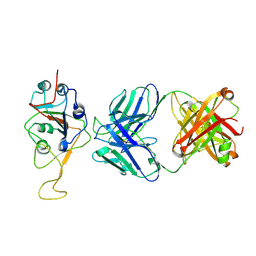 | | Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment (local refinement of the RBD and Fab variable domains) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, SP1-77 Fab heavy chain, SP1-77 Fab light chain, ... | | Authors: | Zhang, J, Luo, S, Kreutzberger, A, Kirchhausen, T, Chen, B, Haynes, B, Alt, F. | | Deposit date: | 2022-04-18 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | An antibody from single human V H -rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion.
Sci Immunol, 7, 2022
|
|
7UPY
 
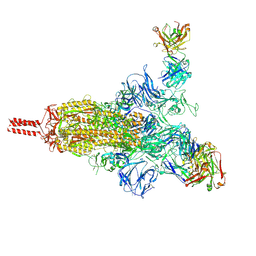 | | An antibody from single human VH-rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Luo, S, Kreutzberger, A, Kirchhausen, T, Chen, B, Haynes, B, Alt, F. | | Deposit date: | 2022-04-18 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | An antibody from single human V H -rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion.
Sci Immunol, 7, 2022
|
|
7UPW
 
 | | Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Luo, S, Kreutzberger, A, Kirchhausen, T, Chen, B, Haynes, B, Alt, F. | | Deposit date: | 2022-04-18 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | An antibody from single human V H -rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion.
Sci Immunol, 7, 2022
|
|
4CK9
 
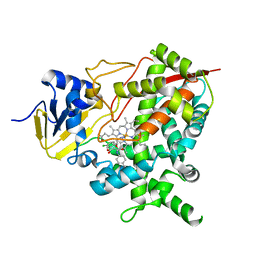 | | STEROL 14-ALPHA DEMETHYLASE (CYP51)FROM TRYPANOSOMA CRUZI IN COMPLEX WITH (S)-1-(4-chlorophenyl)-2-(1H-imidazol-1-yl)ethyl 4- isopropylphenylcarbamate (LFT) | | Descriptor: | (1S)-1-(4-chlorophenyl)-2-(1H-imidazol-1-yl)ethyl [4-(propan-2-yl)phenyl]carbamate, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Friggeri, L, Wawrzak, Z, Tortorella, S, Lepesheva, G.I. | | Deposit date: | 2013-12-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural Basis for Rational Design of Inhibitors Targeting Trypanosoma Cruzi Sterol 14Alpha-Demethylase: Two Regions of the Enzyme Molecule Potentiate its Inhibition.
J.Med.Chem., 57, 2014
|
|
5BUH
 
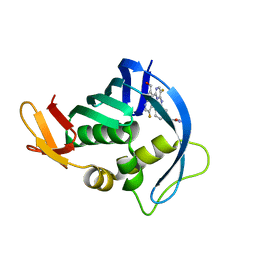 | | Influenza PB2 bound to a hydroxymethyl azaindole inhibitor | | Descriptor: | N-[(1R,3S)-3-({5-fluoro-2-[5-fluoro-2-(hydroxymethyl)-1H-pyrrolo[2,3-b]pyridin-3-yl]pyrimidin-4-yl}amino)cyclohexyl]pyrrolidine-1-carboxamide, Polymerase basic protein 2 | | Authors: | Jacobs, M.D. | | Deposit date: | 2015-06-03 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Novel 2-Substituted 7-Azaindole and 7-Azaindazole Analogues as Potential Antiviral Agents for the Treatment of Influenza.
ACS Med Chem Lett, 8, 2017
|
|
5LN5
 
 | |
5JIG
 
 | | Crytsal structure of Wss1 from S. pombe | | Descriptor: | NICKEL (II) ION, OXYGEN MOLECULE, Ubiquitin and WLM domain-containing metalloprotease SPCC1442.07c | | Authors: | Groll, M, Stingele, J, Boulton, S. | | Deposit date: | 2016-04-22 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Mechanism and Regulation of DNA-Protein Crosslink Repair by the DNA-Dependent Metalloprotease SPRTN.
Mol.Cell, 64, 2016
|
|
6ZLY
 
 | |
6T9C
 
 | |
4RZ2
 
 | |
4RZ3
 
 | | Crystal structure of the MinD-like ATPase FlhG | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Site-determining protein | | Authors: | Schuhmacher, J.S, Bange, G. | | Deposit date: | 2014-12-18 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MinD-like ATPase FlhG effects location and number of bacterial flagella during C-ring assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6G12
 
 | | Crystal structure of GMPPNP bound RbgA from S. aureus | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ribosome biogenesis GTPase A | | Authors: | Pausch, P, Bange, G. | | Deposit date: | 2018-03-20 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.929 Å) | | Cite: | Structural basis for (p)ppGpp-mediated inhibition of the GTPase RbgA.
J. Biol. Chem., 293, 2018
|
|
6G14
 
 | | Crystal structure of ppGpp bound RbgA from S. aureus | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, Ribosome biogenesis GTPase A | | Authors: | Pausch, P, Bange, G. | | Deposit date: | 2018-03-20 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for (p)ppGpp-mediated inhibition of the GTPase RbgA.
J. Biol. Chem., 293, 2018
|
|
6G0Z
 
 | | Crystal structure of GDP bound RbgA from S. aureus | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Ribosome biogenesis GTPase A | | Authors: | Pausch, P, Bange, G. | | Deposit date: | 2018-03-20 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for (p)ppGpp-mediated inhibition of the GTPase RbgA.
J. Biol. Chem., 293, 2018
|
|
6G15
 
 | | Crystal structure of pppGpp bound RbgA from S. aureus | | Descriptor: | Ribosome biogenesis GTPase A, guanosine 5'-(tetrahydrogen triphosphate) 3'-(trihydrogen diphosphate) | | Authors: | Pausch, P, Bange, G. | | Deposit date: | 2018-03-20 | | Release date: | 2018-11-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for (p)ppGpp-mediated inhibition of the GTPase RbgA.
J. Biol. Chem., 293, 2018
|
|
6B8A
 
 | | Crystal structure of MvfR ligand binding domain in complex with M64 | | Descriptor: | 2-[(5-nitro-1H-benzimidazol-2-yl)sulfanyl]-N-(4-phenoxyphenyl)acetamide, COBALT HEXAMMINE(III), DNA-binding transcriptional regulator | | Authors: | Kitao, T, Steinbacher, S, Maskos, K, Blaesse, M, Rahme, L.G. | | Deposit date: | 2017-10-05 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Molecular Insights into Function and Competitive Inhibition ofPseudomonas aeruginosaMultiple Virulence Factor Regulator.
MBio, 9, 2018
|
|
3ZLW
 
 | | Crystal structure of MEK1 in complex with fragment 3 | | Descriptor: | (1R)-1-hydroxy-1-methyl-2,3,6,7-tetrahydro-1H,5H-pyrido[3,2,1-ij]quinolin-5-one, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1 MAPK/ERK KINASE 1, MEK 1, ... | | Authors: | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3ZLX
 
 | | Crystal structure of MEK1 in complex with fragment 18 | | Descriptor: | 7-choro-6-[(3R)-pyrrolidin-3-ylmethoxy]isoquinolin-1(2H)-one, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1 | | Authors: | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3ZLS
 
 | | Crystal structure of MEK1 in complex with fragment 6 | | Descriptor: | 1H-PYRROLO[2,3-B]PYRIDINE-3-CARBOXYLIC ACID, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, SODIUM ION | | Authors: | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3ZM4
 
 | | Crystal structure of MEK1 in complex with fragment 1 | | Descriptor: | 7-chloranyl-6-[(3S)-pyrrolidin-3-yl]oxy-2H-isoquinolin-1-one, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1 | | Authors: | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | Deposit date: | 2013-02-05 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3ZLY
 
 | | Crystal structure of MEK1 in complex with fragment 8 | | Descriptor: | 3-AMINO-1H-INDAZOLE-4-CARBONITRILE, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1, SODIUM ION | | Authors: | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3R8I
 
 | | Crystal Structure of PPARgamma with an achiral ureidofibrate derivative (RT86) | | Descriptor: | 2-(4-{2-[1,3-benzoxazol-2-yl(heptyl)amino]ethyl}phenoxy)-2-methylpropanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Loiodice, F, Laghezza, A, Fracchiolla, G, Lavecchia, A, Novellino, E, Crestani, M. | | Deposit date: | 2011-03-24 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis, characterization and biological evaluation of ureidofibrate-like derivatives endowed with peroxisome proliferator-activated receptor activity.
J.Med.Chem., 55, 2012
|
|
5DEC
 
 | |
4G7G
 
 | | Sterol 14-alpha demethylase (CYP51) from Trypanosoma brucei in complex with the VNI derivative (R)-N-(1-(3,4'-difluorobiphenyl-4-yl)-2-(1H-imidazol-1-yl)ethyl)-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide [VNI/VNF (VFV)] | | Descriptor: | N-[(1R)-1-(3,4'-difluorobiphenyl-4-yl)-2-(1H-imidazol-1-yl)ethyl]-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide, PROTOPORPHYRIN IX CONTAINING FE, sterol 14-alpha-demethylase | | Authors: | Hargrove, T.Y, Wawrzak, Z, Waterman, M.R, Lepesheva, G.I. | | Deposit date: | 2012-07-20 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | VFV as a New Effective CYP51 Structure-Derived Drug Candidate for Chagas Disease and Visceral Leishmaniasis.
J Infect Dis, 212, 2015
|
|
4G3J
 
 | | Sterol 14-alpha demethylase (CYP51) from Trypanosoma brucei in complex with the VNI derivative (R)-N-(1-(2,4-dichlorophenyl)-2-(1H-1,2,4-triazol-1-yl)ethyl)-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide [R-VNI-triazole (VNT)] | | Descriptor: | N-[(1R)-1-(2,4-dichlorophenyl)-2-(1H-1,2,4-triazol-1-yl)ethyl]-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide, PROTOPORPHYRIN IX CONTAINING FE, sterol 14-alpha-demethylase | | Authors: | Hargrove, T.Y, Wawrzak, Z, Waterman, M.R, Lepesheva, G.I. | | Deposit date: | 2012-07-14 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | VFV as a New Effective CYP51 Structure-Derived Drug Candidate for Chagas Disease and Visceral Leishmaniasis.
J Infect Dis, 212, 2015
|
|
