5XHI
 
 | | Crystal structure of Frog M-ferritin D38A mutant | | Descriptor: | CHLORIDE ION, Ferritin, middle subunit, ... | | Authors: | Jagdev, M.K, Vasudevan, D. | | Deposit date: | 2017-04-21 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Surface charge dependent separation of modified and hybrid ferritin in native PAGE: Impact of lysine 104
Biochim. Biophys. Acta, 1865, 2017
|
|
5XHO
 
 | | Crystal structure of Frog M-ferritin E135K mutant | | Descriptor: | CHLORIDE ION, Ferritin, middle subunit, ... | | Authors: | Jagdev, M.K, Vasudevan, D. | | Deposit date: | 2017-04-21 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Surface charge dependent separation of modified and hybrid ferritin in native PAGE: Impact of lysine 104
Biochim. Biophys. Acta, 1865, 2017
|
|
3LCA
 
 | |
1JBK
 
 | |
3KQI
 
 | | crystal structure of PHF2 PHD domain complexed with H3K4Me3 peptide | | Descriptor: | CHLORIDE ION, GLYCEROL, H3K4Me3 peptide, ... | | Authors: | Wen, H, Li, J.Z, Song, T, Lu, M, Lee, M. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-02 | | Last modified: | 2019-02-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Recognition of histone H3K4 trimethylation by the plant homeodomain of PHF2 modulates histone demethylation.
J.Biol.Chem., 285, 2010
|
|
4QQF
 
 | |
3QFP
 
 | | Crystal structure of yeast Hsp70 (Bip/Kar2) ATPase domain | | Descriptor: | 78 kDa glucose-regulated protein homolog, PHOSPHATE ION | | Authors: | Yan, M, Li, J.Z, Sha, B.D. | | Deposit date: | 2011-01-22 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural analysis of the Sil1-Bip complex reveals the mechanism for Sil1 to function as a nucleotide-exchange factor.
Biochem.J., 438, 2011
|
|
3QML
 
 | | The structural analysis of Sil1-Bip complex reveals the mechanism for Sil1 to function as a novel nucleotide exchange factor | | Descriptor: | 78 kDa glucose-regulated protein homolog, MAGNESIUM ION, Nucleotide exchange factor SIL1, ... | | Authors: | Yan, M, Li, J.Z, Sha, B.D. | | Deposit date: | 2011-02-04 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural analysis of the Sil1-Bip complex reveals the mechanism for Sil1 to function as a nucleotide-exchange factor.
Biochem.J., 438, 2011
|
|
3QFU
 
 | | Crystal structure of Yeast Hsp70 (Bip/kar2) complexed with ADP | | Descriptor: | 78 kDa glucose-regulated protein homolog, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yan, M, Li, J.Z, Sha, B.D. | | Deposit date: | 2011-01-22 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the Sil1-Bip complex reveals the mechanism for Sil1 to function as a nucleotide-exchange factor.
Biochem.J., 438, 2011
|
|
3QLE
 
 | | Structural Basis for the Function of Tim50 in the Mitochondrial Presequence Translocase | | Descriptor: | ACETATE ION, CALCIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Qian, X.G, Gebert, M, Hpker, J, Yan, M, Li, J.Z, Wiedemann, N, Laan, M.V.D, Pfanner, N, Sha, B.D. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.831 Å) | | Cite: | Structural basis for the function of tim50 in the mitochondrial presequence translocase.
J.Mol.Biol., 411, 2011
|
|
3QD2
 
 | | Crystal structure of mouse PERK kinase domain | | Descriptor: | Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Wenjun, C, Jingzhi, L, David, R, Bingdong, S. | | Deposit date: | 2011-01-17 | | Release date: | 2011-04-27 | | Last modified: | 2019-01-16 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The structure of the PERK kinase domain suggests the mechanism for its activation.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
6YVX
 
 | | HIF prolyl hydroxylase 2 (PHD2/ EGLN1) in complex with bicyclic BB-287 | | Descriptor: | 4-(isoquinolin-3-ylamino)-4-oxobutanoic acid, BICARBONATE ION, Egl nine homolog 1, ... | | Authors: | Chowdhury, R, Banerji, B, Schofield, C.J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Use of cyclic peptides to induce crystallization: case study with prolyl hydroxylase domain 2.
Sci Rep, 10, 2020
|
|
3H42
 
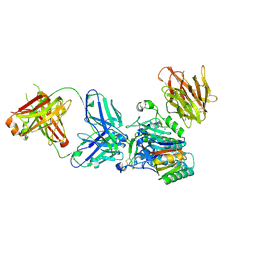 | | Crystal structure of PCSK9 in complex with Fab from LDLR competitive antibody | | Descriptor: | Fab from LDLR competitive antibody: Heavy chain, Fab from LDLR competitive antibody: Light chain, Proprotein convertase subtilisin/kexin type 9, ... | | Authors: | Piper, D.E, Walker, N.P.C, Romanow, W.G, Thibault, S.T, Tsai, M.M, Yang, E. | | Deposit date: | 2009-04-17 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | From the Cover: A proprotein convertase subtilisin/kexin type 9 neutralizing antibody reduces serum cholesterol in mice and nonhuman primates.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6YW4
 
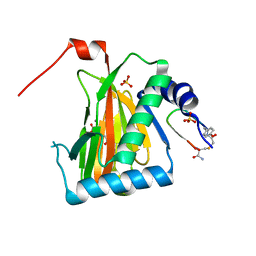 | |
6UUM
 
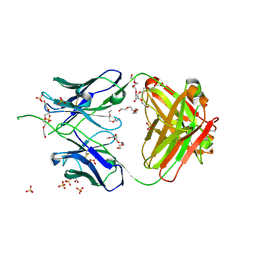 | | Crystal structure of antibody 438-B11 DSS mutant (Cys98A-Cys100aA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, B11 DSS Fab Heavy Chain, ... | | Authors: | Kumar, S, Wilson, I.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A V H 1-69 antibody lineage from an infected Chinese donor potently neutralizes HIV-1 by targeting the V3 glycan supersite.
Sci Adv, 6, 2020
|
|
6UUL
 
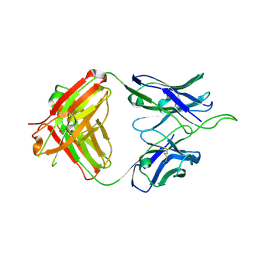 | | Crystal structure of broad and potent HIV-1 neutralizing antibody 438-D5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, D5 Fab Heavy Chain, ... | | Authors: | Kumar, S, Wilson, I.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | A V H 1-69 antibody lineage from an infected Chinese donor potently neutralizes HIV-1 by targeting the V3 glycan supersite.
Sci Adv, 6, 2020
|
|
6UTK
 
 | |
6UUH
 
 | |
6V6W
 
 | |
5K0M
 
 | | Targeting the PRC2 complex through a novel protein-protein interaction inhibitor of EED | | Descriptor: | (3R,4S)-1-[(1S)-7-fluoro-2,3-dihydro-1H-inden-1-yl]-N,N-dimethyl-4-{4-[4-(methylsulfonyl)piperazin-1-yl]phenyl}pyrrolidin-3-amine, Polycomb protein EED | | Authors: | Jakob, C.G, Bigelow, L.J, Zhu, H, Sun, C. | | Deposit date: | 2016-05-17 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The EED protein-protein interaction inhibitor A-395 inactivates the PRC2 complex.
Nat. Chem. Biol., 13, 2017
|
|
5KJ2
 
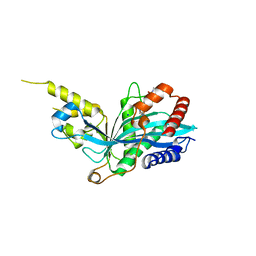 | | The novel p300/CBP inhibitor A-485 uncovers a unique mechanism of action to target AR in castrate resistant prostate cancer | | Descriptor: | Histone acetyltransferase p300, N-[(4-fluorophenyl)methyl]-2-{(1R)-5-[(methylcarbamoyl)amino]-2',4'-dioxo-2,3-dihydro-3'H-spiro[indene-1,5'-[1,3]oxazolidin]-3'-yl}-N-[(2S)-1,1,1-trifluoropropan-2-yl]acetamide, SODIUM ION | | Authors: | Jakob, C.G, Qiu, W, Edalji, R.P, Sun, C. | | Deposit date: | 2016-06-17 | | Release date: | 2017-09-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours.
Nature, 550, 2017
|
|
4UF1
 
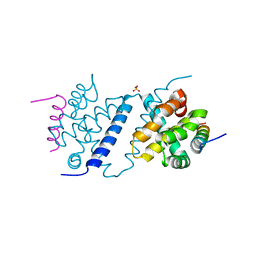 | | Deerpox virus DPV022 in complex with Bak BH3 | | Descriptor: | Antiapoptotic membrane protein, Bcl-2 homologous antagonist/killer, SULFATE ION | | Authors: | Burton, D.R, Kvansakul, M. | | Deposit date: | 2014-12-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Deerpox Virus-Mediated Inhibition of Apoptosis.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UF2
 
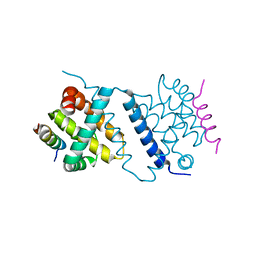 | | Deerpox virus DPV022 in complex with Bax BH3 | | Descriptor: | Antiapoptotic membrane protein, Apoptosis regulator BAX | | Authors: | Burton, D.R, Kvansakul, M. | | Deposit date: | 2014-12-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of Deerpox Virus-Mediated Inhibition of Apoptosis.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UF3
 
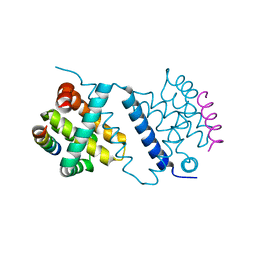 | | Deerpox virus DPV022 in complex with Bim BH3 | | Descriptor: | Antiapoptotic membrane protein, Bcl-2-like protein 11 | | Authors: | Burton, D.R, Kvansakul, M. | | Deposit date: | 2014-12-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Deerpox Virus-Mediated Inhibition of Apoptosis.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8CK2
 
 | | MUC2 CysD1 G1352S | | Descriptor: | ACETATE ION, CALCIUM ION, Mucin-2, ... | | Authors: | Khmelnitsky, L, Fass, D. | | Deposit date: | 2023-02-14 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Pulmonary Fibrosis Mutation Undermines Mucin 5B Supramolecular Assembly
To Be Published
|
|
