6A4D
 
 | |
6A4E
 
 | |
6A7I
 
 | | CYP154C4 from Streptomyces sp. W2061 | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, C.W, Lee, J.H. | | Deposit date: | 2018-07-03 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Characterization of two steroid hydroxylases from different Streptomyces spp. and their ligand-bound and -unbound crystal structures.
Febs J., 286, 2019
|
|
6A4A
 
 | |
6A4F
 
 | |
6AGQ
 
 | | Acetyl xylan esterase from Paenibacillus sp. R4 | | Descriptor: | ZINC ION, acetyl xylan esterase | | Authors: | Park, S, Lee, C.W, Lee, J.H. | | Deposit date: | 2018-08-13 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and functional characterization of a cold-active acetyl xylan esterase (PbAcE) from psychrophilic soil microbe Paenibacillus sp.
PLoS ONE, 13, 2018
|
|
8GTL
 
 | | Crystal Structure of Cytochrome P450 (CYP101D5) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450 CYP101D5 | | Authors: | Do, H, Lee, J.H. | | Deposit date: | 2022-09-08 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure and Biochemical Analysis of a Cytochrome P450 CYP101D5 from Sphingomonas echinoides.
Int J Mol Sci, 23, 2022
|
|
8HEA
 
 | | Esterase2 (EaEst2) from Exiguobacterium antarcticum | | Descriptor: | Thermostable carboxylesterase Est30 | | Authors: | Hwang, J, Lee, J.H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural and Biochemical Insights into Bis(2-hydroxyethyl) Terephthalate Degrading Carboxylesterase Isolated from Psychrotrophic Bacterium Exiguobacterium antarcticum.
Int J Mol Sci, 24, 2023
|
|
8ILJ
 
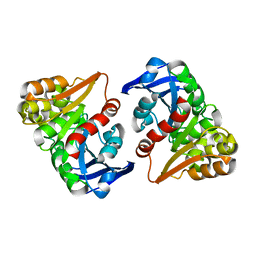 | |
8JUO
 
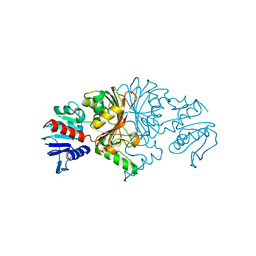 | |
8JUS
 
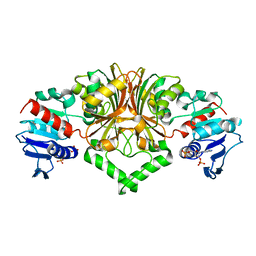 | |
6X6P
 
 | | Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Herrera, N.G, Morano, N.C, Celikgil, A, Georgiev, G.I, Malonis, R, Lee, J.H, Tong, K, Vergnolle, O, Massimi, A, Yen, L.Y, Noble, A.J, Kopylov, M, Bonanno, J.B, Garrett-Thompson, S.C, Hayes, D.B, Brenowitz, M, Garforth, S.J, Eng, E.T, Lai, J.R, Almo, S.C. | | Deposit date: | 2020-05-28 | | Release date: | 2020-06-10 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis.
Biorxiv, 2020
|
|
5XNT
 
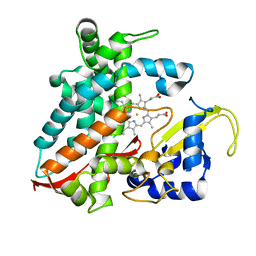 | | Structure of CYP106A2 from Bacillus sp. PAMC 23377 | | Descriptor: | Cytochrome P450 CYP106, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, C.W, Kim, K.-H, Bikash, D, Park, S.-H, Park, H, Oh, T.-J, Lee, J.H. | | Deposit date: | 2017-05-24 | | Release date: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure and Functional Characterization of a Cytochrome P450 (BaCYP106A2) fromBacillussp. PAMC 23377.
J. Microbiol. Biotechnol., 27, 2017
|
|
6LVO
 
 | |
6LVP
 
 | |
6L3A
 
 | | Cytochrome P450 107G1 (RapN) with everolimus | | Descriptor: | Cytochrome P450, Everolimus, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Km, V.C, Kim, D.H, Lim, Y.R, Lee, I.H, Lee, J.H, Kang, L.W. | | Deposit date: | 2019-10-10 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into CYP107G1 from rapamycin-producing Streptomyces rapamycinicus.
Arch.Biochem.Biophys., 692, 2020
|
|
6L39
 
 | | Cytochrome P450 107G1 (RapN) | | Descriptor: | Cytochrome P450, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Kim, V.C, Kim, D.H, Lim, Y.R, Lee, I.H, Lee, J.H, Kang, L.W. | | Deposit date: | 2019-10-10 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural insights into CYP107G1 from rapamycin-producing Streptomyces rapamycinicus.
Arch.Biochem.Biophys., 692, 2020
|
|
8GJE
 
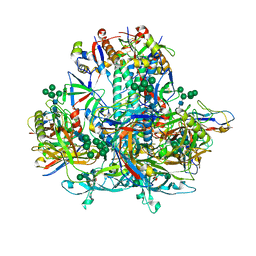 | | HIV-1 Env subtype C CZA97.12 SOSIP.664 in complex with 3BNC117 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3BNC117 Fab heavy chain, ... | | Authors: | Ozorowski, G, Lee, J.H, Ward, A.B. | | Deposit date: | 2023-03-15 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Glycan heterogeneity as a cause of the persistent fraction in HIV-1 neutralization.
Plos Pathog., 19, 2023
|
|
6J4N
 
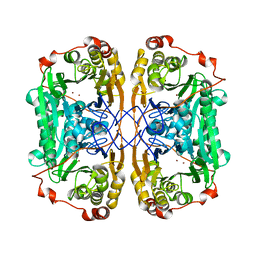 | | Structure of papua new guinea MBL-1(PNGM-1) native | | Descriptor: | Metallo-beta-lactamases PNGM-1, ZINC ION | | Authors: | Hong, M.K, Park, K.S, Jeon, J.H, Lee, J.H, Park, Y.S, Lee, S.H, Kang, L.W. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The novel metallo-beta-lactamase PNGM-1 from a deep-sea sediment metagenome: crystallization and X-ray crystallographic analysis.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
4NU2
 
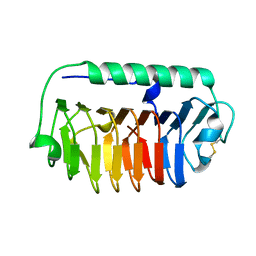 | | Crystal structure of an ice-binding protein (FfIBP) from the Antarctic bacterium, Flavobacterium frigoris PS1 | | Descriptor: | Antifreeze protein | | Authors: | Do, H, Kim, S.J, Lee, S.G, Park, H, Kim, H.J, Lee, J.H. | | Deposit date: | 2013-12-03 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based characterization and antifreeze properties of a hyperactive ice-binding protein from the Antarctic bacterium Flavobacterium frigoris PS1
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NUH
 
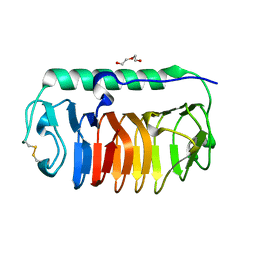 | | Crystal structure of mLeIBP, a capping head region swapped mutant of ice-binding protein | | Descriptor: | DI(HYDROXYETHYL)ETHER, ice-binding protein | | Authors: | Do, H, Kim, S.J, Lee, S.G, Park, H, Kim, H.J, Lee, J.H. | | Deposit date: | 2013-12-03 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structure-based characterization and antifreeze properties of a hyperactive ice-binding protein from the Antarctic bacterium Flavobacterium frigoris PS1
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8ISP
 
 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by cephalexin | | Descriptor: | (R)-2-((R)-((R)-2-amino-2-phenylacetamido)(carboxy)methyl)-5-methyl-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
8ISO
 
 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Beta-lactamase | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
8ISQ
 
 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, ... | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
8ISR
 
 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by cefaclor | | Descriptor: | (R)-2-((R)-((R)-2-amino-2-phenylacetamido)(carboxy)methyl)-5-chloro-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
