3DL7
 
 | | Aged Form of Mouse Acetylcholinesterase Inhibited by Tabun- Update | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, CHLORIDE ION, ... | | Authors: | Carletti, E, Li, H, Li, B, Ekstrom, F, Nicolet, Y, Loiodice, M, Gillon, E, Froment, M.T, Lockridge, O, Schopfer, L.M, Masson, P, Nachon, F. | | Deposit date: | 2008-06-26 | | Release date: | 2008-12-02 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Aging of Cholinesterases Phosphylated by Tabun Proceeds through O-Dealkylation.
J.Am.Chem.Soc., 130, 2008
|
|
5JJY
 
 | | Crystal structure of SETD2 bound to histone H3.3 K36M peptide | | Descriptor: | Histone H3.3, Histone-lysine N-methyltransferase SETD2, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Yang, S, Zheng, X, Li, H. | | Deposit date: | 2016-04-25 | | Release date: | 2016-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Molecular basis for oncohistone H3 recognition by SETD2 methyltransferase
Genes Dev., 30, 2016
|
|
3OHN
 
 | | Crystal structure of the FimD translocation domain | | Descriptor: | Outer membrane usher protein FimD | | Authors: | Wang, T, Li, H. | | Deposit date: | 2010-08-17 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Crystal structure of the FimD usher bound to its cognate FimC-FimH substrate.
Nature, 474, 2011
|
|
3N6F
 
 | | Structure of endothelial nitric oxide synthase N368D single mutant heme domain complexed with 6,6'-(2,2'-(pyridine-3,5-diyl)bis(ethane-2,1-diyl))bis(4-methylpyridin-2-amine) | | Descriptor: | 'Nitric oxide synthase, 5,6,7,8-TETRAHYDROBIOPTERIN, 6,6'-(pyridine-3,5-diyldiethane-2,1-diyl)bis(4-methylpyridin-2-amine), ... | | Authors: | Delker, S.L, Li, H, Poulos, T.L. | | Deposit date: | 2010-05-25 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Role of zinc in isoform-selective inhibitor binding to neuronal nitric oxide synthase .
Biochemistry, 49, 2010
|
|
3N6C
 
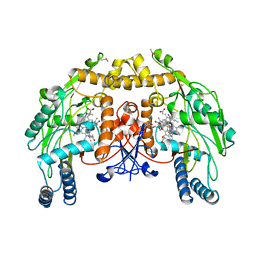 | | Structure of endothelial nitric oxide synthase H373S single mutant heme domain complexed with 4-(2-(6-(2-(6-amino-4-methylpyridin-2-yl)ethyl)pyridin-2-yl)ethyl)-6-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{6-[2-(2-amino-6-methylpyridin-4-yl)ethyl]pyridin-2-yl}ethyl)-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Delker, S.L, Li, H, Poulos, T.L. | | Deposit date: | 2010-05-25 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Role of zinc in isoform-selective inhibitor binding to neuronal nitric oxide synthase .
Biochemistry, 49, 2010
|
|
3N5T
 
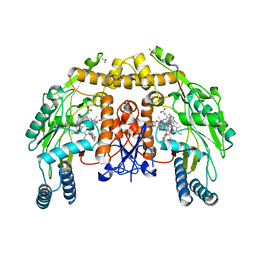 | | Structure of endothelial nitric oxide synthase heme domain complexed with 6,6'-(2,2'-(pyridine-3,5-diyl)bis(ethane-2,1-diyl))bis(4-methylpyridin-2-amine) | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6,6'-(pyridine-3,5-diyldiethane-2,1-diyl)bis(4-methylpyridin-2-amine), ACETATE ION, ... | | Authors: | Delker, S.L, Li, H, Poulos, T.L. | | Deposit date: | 2010-05-25 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Role of zinc in isoform-selective inhibitor binding to neuronal nitric oxide synthase .
Biochemistry, 49, 2010
|
|
3N6B
 
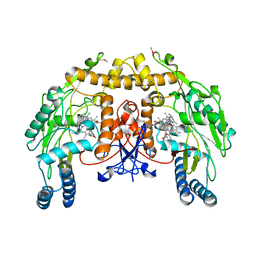 | | Structure of endothelial nitric oxide synthase H373S single mutant heme domain complexed with 6,6'-(2,2'-(pyridine-3,5-diyl)bis(ethane-2,1-diyl))bis(4-methylpyridin-2-amine) | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6,6'-(pyridine-3,5-diyldiethane-2,1-diyl)bis(4-methylpyridin-2-amine), CACODYLIC ACID, ... | | Authors: | Delker, S.L, Li, H, Poulos, T.L. | | Deposit date: | 2010-05-25 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Role of zinc in isoform-selective inhibitor binding to neuronal nitric oxide synthase .
Biochemistry, 49, 2010
|
|
3N68
 
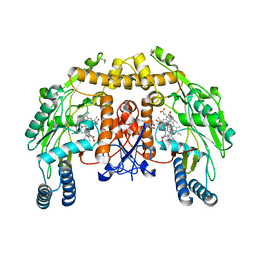 | | Structure of endothelial nitric oxide synthase heme domain N368D/V106M double mutant complexed with 4-(3-(2-(6-amino-4-methylpyridin-2-yl)ethyl)phenethyl)-6-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{3-[2-(2-amino-6-methylpyridin-4-yl)ethyl]phenyl}ethyl)-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Delker, S.L, Li, H, Poulos, T.L. | | Deposit date: | 2010-05-25 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Role of zinc in isoform-selective inhibitor binding to neuronal nitric oxide synthase .
Biochemistry, 49, 2010
|
|
3ER8
 
 | | Crystal structure of the heterodimeric vaccinia virus mRNA polyadenylate polymerase complex with two fragments of RNA | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase, Poly(A) polymerase catalytic subunit, RNA/DNA chimera 5'-D(CP*CP*)R(UP*UP*)D(C)-3', ... | | Authors: | Li, C, Li, H, Zhou, S, Poulos, T.L, Gershon, P.D. | | Deposit date: | 2008-10-01 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Polymerase Translocation with Respect to Single-Stranded Nucleic Acid: Looping or Wrapping of Primer around a Poly(A) Polymerase
Structure, 17, 2009
|
|
5VVX
 
 | | Structural Investigations of the Substrate Specificity of Human O-GlcNAcase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lamin B1, Protein O-GlcNAcase | | Authors: | Li, B, Jiang, J, Li, H, Hu, C.-W. | | Deposit date: | 2017-05-21 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the substrate binding adaptability and specificity of human O-GlcNAcase.
Nat Commun, 8, 2017
|
|
3DL4
 
 | | Non-Aged Form of Mouse Acetylcholinesterase Inhibited by Tabun- Update | | Descriptor: | Acetylcholinesterase, HEXAETHYLENE GLYCOL | | Authors: | Carletti, E, Li, H, Li, B, Ekstrom, F, Nicolet, Y, Loiodice, M, Gillon, E, Froment, M.T, Lockridge, O, Schopfer, L.M, Masson, P, Nachon, F. | | Deposit date: | 2008-06-26 | | Release date: | 2008-12-02 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Aging of Cholinesterases Phosphylated by Tabun Proceeds through O-Dealkylation.
J.Am.Chem.Soc., 130, 2008
|
|
5VVO
 
 | |
5VVU
 
 | | Structural Investigations of the Substrate Specificity of Human O-GlcNAcase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein O-GlcNAcase, TAB1 peptide | | Authors: | Li, B, Jiang, J, Li, H, Hu, C.-W. | | Deposit date: | 2017-05-20 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the substrate binding adaptability and specificity of human O-GlcNAcase.
Nat Commun, 8, 2017
|
|
1QR5
 
 | | SOLUTION STRUCTURE OF HISTIDINE CONTAINING PROTEIN (HPR) FROM STAPHYLOCOCCUS CARNOSUS | | Descriptor: | PHOSPHOCARRIER PROTEIN HPR | | Authors: | Kalbitzer, H.R, Gorler, A, Li, H, Dubovskii, P.V, Hengstenberg, W, Kowolik, C, Yamada, H, Akasaka, K. | | Deposit date: | 1999-05-19 | | Release date: | 2000-06-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | 15N and 1H NMR study of histidine containing protein (HPr) from Staphylococcus carnosus at high pressure.
Protein Sci., 9, 2000
|
|
3N5Q
 
 | | Structure of endothelial nitric oxide synthase heme domain complexed with 6,6'-(2,2'-(5-amino-1,3-phenylene)bis(ethane-2,1-diyl))bis(4-methylpyridin-2-amine) | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6,6'-[(5-aminobenzene-1,3-diyl)diethane-2,1-diyl]bis(4-methylpyridin-2-amine), CACODYLIC ACID, ... | | Authors: | Delker, S.L, Li, H, Poulos, T.L. | | Deposit date: | 2010-05-25 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Role of zinc in isoform-selective inhibitor binding to neuronal nitric oxide synthase .
Biochemistry, 49, 2010
|
|
3N67
 
 | | Structure of endothelial nitric oxide synthase N368D/V106M double mutant heme domain complexed with 6,6'-(2,2'-(5-amino-1,3-phenylene)bis(ethane-2,1-diyl))bis(4-methylpyridin-2-amine) | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6,6'-[(5-aminobenzene-1,3-diyl)diethane-2,1-diyl]bis(4-methylpyridin-2-amine), ACETATE ION, ... | | Authors: | Delker, S.L, Li, H, Poulos, T.L. | | Deposit date: | 2010-05-25 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Role of zinc in isoform-selective inhibitor binding to neuronal nitric oxide synthase .
Biochemistry, 49, 2010
|
|
5VVT
 
 | | Structural Investigations of the Substrate Specificity of Human O-GlcNAcase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ELK1 peptide, Protein O-GlcNAcase | | Authors: | Li, B, Jiang, J, Li, H, Hu, C.-W. | | Deposit date: | 2017-05-20 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the substrate binding adaptability and specificity of human O-GlcNAcase.
Nat Commun, 8, 2017
|
|
5VVV
 
 | | Structural Investigations of the Substrate Specificity of Human O-GlcNAcase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein O-GlcNAcase, a-crystallin B | | Authors: | Li, B, Jiang, J, Li, H, Hu, C.-W. | | Deposit date: | 2017-05-20 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the substrate binding adaptability and specificity of human O-GlcNAcase.
Nat Commun, 8, 2017
|
|
3PNH
 
 | | Structure of Bovine Endothelial Nitric Oxide Synthase Heme Domain in complex with 6-(((3R,4R)-4-(2-((2-FLUORO-2-(3-FLUOROPHENYL) ETHYL)AMINO)ETHOXY)PYRROLIDIN-3-YL)METHYL)-4-METHYLPYRIDIN-2-AMINE | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3R,4R)-4-(2-{[(2R)-2-fluoro-2-(3-fluorophenyl)ethyl]amino}ethoxy)pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Delker, S.L, Li, H, Poulos, T.L. | | Deposit date: | 2010-11-18 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Improved Synthesis of Chiral Pyrrolidine Inhibitors and Their Binding Properties to Neuronal Nitric Oxide Synthase.
J.Med.Chem., 54, 2011
|
|
3ERC
 
 | | Crystal structure of the heterodimeric vaccinia virus mRNA polyadenylate polymerase with three fragments of RNA and 3'-deoxy ATP | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase, ... | | Authors: | Li, C, Li, H, Zhou, S, Poulos, T.L, Gershon, P.D. | | Deposit date: | 2008-10-01 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Polymerase Translocation with Respect to Single-Stranded Nucleic Acid: Looping or Wrapping of Primer around a Poly(A) Polymerase
Structure, 17, 2009
|
|
7SH2
 
 | | Structure of the yeast Rad24-RFC loader bound to DNA and the open 9-1-1 clamp | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, Crick strand, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2021-10-07 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | DNA is loaded through the 9-1-1 DNA checkpoint clamp in the opposite direction of the PCNA clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SGZ
 
 | | Structure of the yeast Rad24-RFC loader bound to DNA and the closed 9-1-1 clamp | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, Crick strand, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2021-10-07 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | DNA is loaded through the 9-1-1 DNA checkpoint clamp in the opposite direction of the PCNA clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5TRR
 
 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2169 | | Descriptor: | N,N-diethyl-N~2~-(3-phenylpropanoyl)-L-asparaginyl-N-[(naphthalen-1-yl)methyl]-L-alaninamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
5TRG
 
 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide DPLG-2 | | Descriptor: | N,N-diethyl-N~2~-[(2E)-3-phenylprop-2-enoyl]-L-asparaginyl-4-fluoro-N-[(naphthalen-1-yl)methyl]-L-phenylalaninamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, R.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-26 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
6AYF
 
 | | TRPML3/ML-SA1 complex at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Mucolipin-3 | | Authors: | Zhou, X, Li, M, Su, D, Jia, Q, Li, H, Li, X, Yang, J. | | Deposit date: | 2017-09-08 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Cryo-EM structures of the human endolysosomal TRPML3 channel in three distinct states.
Nat. Struct. Mol. Biol., 24, 2017
|
|
