9I02
 
 | | Structure of recombinant human butyrylcholinesterase in complex with (S)-N-((1-benzylpyrrolidin-3-yl)methyl)-N-methylnaphthalene-2-sulfonamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Kosak, U, Gobec, S, Nachon, F. | | Deposit date: | 2025-01-14 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Lead Optimization of a Butyrylcholinesterase Inhibitor for the Treatment of Alzheimer's Disease.
J.Med.Chem., 68, 2025
|
|
9I03
 
 | | Structure of recombinant human butyrylcholinesterase in complex with (R)-N-((1-benzylpyrrolidin-3-yl)methyl)-N-methylnaphthalene-2-sulfonamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Kosak, U, Gobec, S, Nachon, F. | | Deposit date: | 2025-01-14 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Lead Optimization of a Butyrylcholinesterase Inhibitor for the Treatment of Alzheimer's Disease.
J.Med.Chem., 68, 2025
|
|
6KHY
 
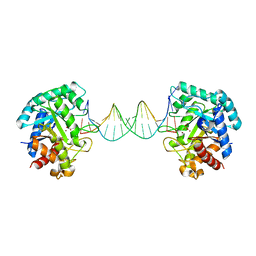 | | The crystal structure of AsfvAP:AG | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (AGCGTCACCGACGAGGC), DNA(AGCGTCACCGACGAGG), ... | | Authors: | Chen, Y.Q, Gan, J.H. | | Deposit date: | 2019-07-16 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.008 Å) | | Cite: | A unique DNA-binding mode of African swine fever virus AP endonuclease.
Cell Discov, 6, 2020
|
|
9IYQ
 
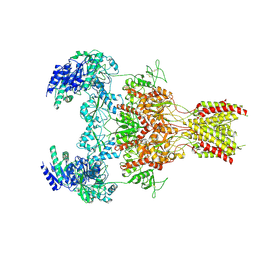 | | Structure of the human GluN1-N2B NMDA receptors in the Mg2+ free state | | Descriptor: | (2R)-4-(3-phosphonopropyl)piperazine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Huang, X, Sun, X, Zhu, S. | | Deposit date: | 2024-07-31 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural insights into the diverse actions of magnesium on NMDA receptors.
Neuron, 113, 2025
|
|
9IYP
 
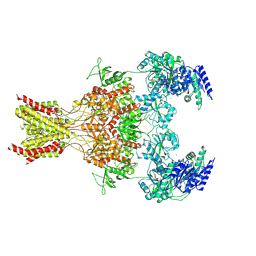 | | Structure of the human GluN1-N2B NMDA receptors in the Mg2+ bound state | | Descriptor: | (2R)-4-(3-phosphonopropyl)piperazine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Huang, X, Sun, X, Zhu, S. | | Deposit date: | 2024-07-31 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural insights into the diverse actions of magnesium on NMDA receptors.
Neuron, 113, 2025
|
|
5X4R
 
 | | Structure of the N-terminal domain (NTD) of MERS-CoV spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
9J3C
 
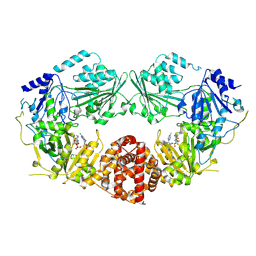 | | Cryo-EM structure of NAT10 with Co-enzyme A | | Descriptor: | COENZYME A, RNA cytidine acetyltransferase | | Authors: | Jiang, Y, Xia, J. | | Deposit date: | 2024-08-08 | | Release date: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Targeting NAT10 attenuates homologous recombination via destabilizing DNA:RNA hybrids and overcomes PARP inhibitor resistance in cancers.
Drug Resist Updat., 81, 2025
|
|
6K5L
 
 | | The crystal structure of isocitrate dehydrogenase kinase/phosphatase wtih two Mn2+ from E. coli | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, Isocitrate dehydrogenase kinase/phosphatase, ... | | Authors: | Zhang, X, Lei, Z, Zheng, J, Jia, Z. | | Deposit date: | 2019-05-29 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Characterization of metal binding of bifunctional kinase/phosphatase AceK and implication in activity modulation.
Sci Rep, 9, 2019
|
|
6KI3
 
 | | The crystal structure of AsfvAP:dF commplex | | Descriptor: | DNA (5'-D(*CP*CP*TP*CP*GP*TP*CP*GP*GP*GP*GP*AP*CP*GP*CP*TP*G)-3'), DNA (5'-D(*GP*CP*AP*GP*CP*GP*TP*CP*C)-3'), DNA (5'-D(P*(3DR)P*CP*GP*AP*CP*GP*AP*G)-3'), ... | | Authors: | Chen, Y, Gan, J. | | Deposit date: | 2019-07-17 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.354 Å) | | Cite: | A unique DNA-binding mode of African swine fever virus AP endonuclease.
Cell Discov, 6, 2020
|
|
7DHX
 
 | | Crystal structure of SARS-CoV-2 RBD binding to pangolin ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ZINC ION, ... | | Authors: | Wang, Q.H, Qi, J.X, Wu, L.L. | | Deposit date: | 2020-11-17 | | Release date: | 2021-09-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of pangolin ACE2 engaged by COVID-19 virus
Chin.Sci.Bull., 66, 2021
|
|
7CUT
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with Z-VAD-FMK | | Descriptor: | 3C protein, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Lu, M, Yang, H.T, Wang, Z.Y, Zhao, Y, Xing, Y.F. | | Deposit date: | 2020-08-24 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Identification of proteasome and caspase inhibitors targeting SARS-CoV-2 M pro .
Signal Transduct Target Ther, 6, 2021
|
|
7CUU
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with MG132 | | Descriptor: | 3C protein, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Lu, M, Yang, H.T, Wang, Z.Y, Zhao, Y, Xing, Y.F. | | Deposit date: | 2020-08-24 | | Release date: | 2021-06-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Identification of proteasome and caspase inhibitors targeting SARS-CoV-2 M pro .
Signal Transduct Target Ther, 6, 2021
|
|
7EIN
 
 | | SARS-CoV-2 main proteinase complex with microbial metabolite leupeptin | | Descriptor: | 3C-like proteinase, leupeptin | | Authors: | Fu, L.F, Feng, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-03-31 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Microbial Metabolite Leupeptin in the Treatment of COVID-19 by Traditional Chinese Medicine Herbs.
Mbio, 12, 2021
|
|
4AV4
 
 | | FimH lectin domain co-crystal with a alpha-D-mannoside O-linked to a propynyl pyridine | | Descriptor: | 3-(pyridin-3-yl)prop-2-yn-1-yl alpha-D-mannopyranoside, FIMH | | Authors: | Bouckaert, J, Touaibia, M, Roos, G, Shiao, T.C, Wang, Q, Papadopoulos, A, Roy, R. | | Deposit date: | 2012-05-23 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Tyrosine Gate as a Potential Entropic Lever in the Receptor-Binding Site of the Bacterial Adhesin Fimh.
Biochemistry, 51, 2012
|
|
4AUJ
 
 | | FimH lectin domain co-crystal with a alpha-D-mannoside O-linked to para hydroxypropargyl phenyl | | Descriptor: | 4-(3-hydroxyprop-1-yn-1-yl)phenyl alpha-D-mannopyranoside, FIMH | | Authors: | Bouckaert, J, Touaibia, M, Roos, G, Shiao, T.C, Wang, Q, Papadopoulos, A, Roy, R. | | Deposit date: | 2012-05-17 | | Release date: | 2013-05-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.527 Å) | | Cite: | Validation of Reactivity Descriptors to Assess the Aromatic Stacking within the Tyrosine Gate of Fimh.
Acs Med.Chem.Lett., 4, 2013
|
|
7WE6
 
 | | Structure of Csy-AcrIF24-dsDNA | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | Authors: | Zhang, L, Feng, Y. | | Deposit date: | 2021-12-22 | | Release date: | 2022-04-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24.
Nat Commun, 13, 2022
|
|
4ATT
 
 | | FimH lectin domain co-crystal with a alpha-D-mannoside O-linked to a propynyl para methoxy phenyl | | Descriptor: | 3-(4-methoxyphenyl)prop-2-yn-1-yl alpha-D-mannopyranoside, FIMH | | Authors: | Bouckaert, J, Touaibia, M, Roos, G, Shiao, T.C, Wang, Q, Papadopoulos, A, Roy, R. | | Deposit date: | 2012-05-09 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Validation of Reactivity Descriptors to Assess the Aromatic Stacking within the Tyrosine Gate of Fimh.
Acs Med.Chem.Lett., 4, 2013
|
|
7XA7
 
 | | Crystal structure of SARS-CoV-2 receptor-binding domain in complex with intermediate horseshoe bat ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Tang, L.F, Zhang, D, Han, P, Qi, J.X. | | Deposit date: | 2022-03-17 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structural basis of SARS-CoV-2 and its variants binding to intermediate horseshoe bat ACE2.
Int J Biol Sci, 18, 2022
|
|
7X8Q
 
 | | Frizzled-10 CRD in complex with F10_A9 Fab | | Descriptor: | Antibody F10_A9 Fab, Heavy chain, Light chain, ... | | Authors: | Ge, Q, Wang, Q. | | Deposit date: | 2022-03-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | An epitope-directed selection strategy facilitating the identification of Frizzled receptor selective antibodies.
Structure, 31, 2023
|
|
7X8P
 
 | | Frizzled 2 CRD in complex with pF7_A5 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody pF7_A5 Fab, Heavy chain, ... | | Authors: | Ge, Q, Wang, Q. | | Deposit date: | 2022-03-14 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | An epitope-directed selection strategy facilitating the identification of Frizzled receptor selective antibodies.
Structure, 31, 2023
|
|
7X8T
 
 | | Frizzled 10 CRD in complex with hB9L9.3 Fab | | Descriptor: | Antibody hB9L9.3 Fab, Heavy chain, Light chain, ... | | Authors: | Ge, Q, Wang, Q. | | Deposit date: | 2022-03-14 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | An epitope-directed selection strategy facilitating the identification of Frizzled receptor selective antibodies.
Structure, 31, 2023
|
|
8VFX
 
 | | Cryo-EM structure of 186bp ALBN1 nucleosome aided by scFv | | Descriptor: | DNA (158-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Zhou, B.R, Bai, Y. | | Deposit date: | 2023-12-22 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structural insights into the cooperative nucleosome recognition and chromatin opening by FOXA1 and GATA4.
Mol.Cell, 84, 2024
|
|
8VFY
 
 | |
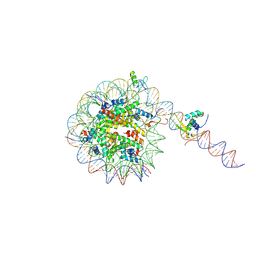
8VG1
 
 | |
8VG0
 
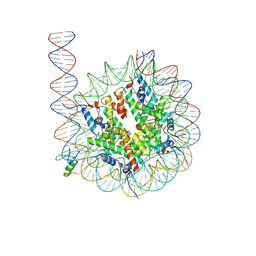 | | Cryo-EM structure of GATA4 in complex with ALBN1 nucleosome | | Descriptor: | DNA (159-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Zhou, B.R, Bai, Y. | | Deposit date: | 2023-12-22 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural insights into the cooperative nucleosome recognition and chromatin opening by FOXA1 and GATA4.
Mol.Cell, 84, 2024
|
|
