3SVI
 
 | | Structure of the Pto-binding domain of HopPmaL generated by limited thermolysin digestion | | Descriptor: | CHLORIDE ION, SODIUM ION, SULFATE ION, ... | | Authors: | Singer, A.U, Stein, A, Xu, X, Cui, H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-07-12 | | Release date: | 2011-08-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of HopPmaL Reveals the Presence of a Second Adaptor Domain Common to the HopAB Family of Pseudomonas syringae Type III Effectors.
Biochemistry, 51, 2012
|
|
3TJY
 
 | | Structure of the Pto-binding domain of HopPmaL generated by limited chymotrypsin digestion | | Descriptor: | CHLORIDE ION, Effector protein hopAB3, SULFATE ION | | Authors: | Singer, A.U, Stein, A, Xu, X, Cui, H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-08-25 | | Release date: | 2011-09-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of HopPmaL reveals the presence of a second adaptor domain common to the HopAB family of Pseudomonas syringae type III effectors.
Biochemistry, 51, 2012
|
|
3I4P
 
 | | Crystal structure of AsnC family transcriptional regulator from Agrobacterium tumefaciens | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Cymborowski, M, Chruszcz, M, Luo, H, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-02 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of AsnC family transcriptional regulator from Agrobacterium tumefaciens
To be Published
|
|
3EA0
 
 | | Crystal Structure of ParA Family ATPase from Chlorobium tepidum TLS | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase, ParA family, ... | | Authors: | Kim, Y, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-08-24 | | Release date: | 2008-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of ParA Family ATPase from Chlorobium tepidum TLS
To be Published
|
|
3EAG
 
 | |
3IC5
 
 | |
3IC9
 
 | | The structure of dihydrolipoamide dehydrogenase from Colwellia psychrerythraea 34H. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SODIUM ION, dihydrolipoamide dehydrogenase | | Authors: | Tan, K, Rakowski, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-17 | | Release date: | 2009-07-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of dihydrolipoamide dehydrogenase from Colwellia psychrerythraea 34H.
To be Published
|
|
3IR9
 
 | |
3IX7
 
 | |
3IC6
 
 | |
3IRA
 
 | |
3S9X
 
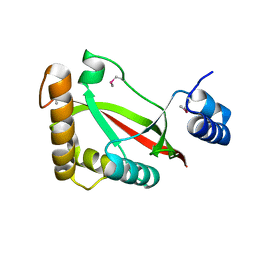 | | High resolution crystal structure of ASCH domain from Lactobacillus crispatus JV V101 | | Descriptor: | ASCH domain, CHLORIDE ION | | Authors: | Nocek, B, Xu, X, Cui, H, Jedrzejczak, R, Edwards, A, Savchenko, A, Mabbutt, B.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-06-02 | | Release date: | 2011-07-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High resolution crystal structure of ASCH domain from Lactobacillus crispatus JV V101
TO BE PUBLISHED
|
|
3I8N
 
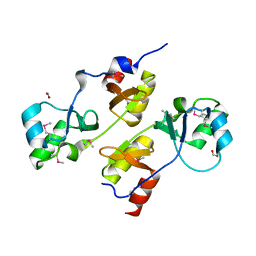 | |
3ELK
 
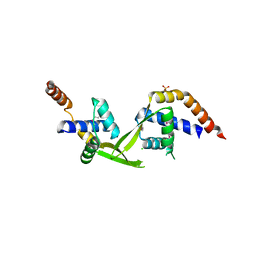 | | Crystal structure of putative transcriptional regulator TA0346 from Thermoplasma acidophilum | | Descriptor: | CHLORIDE ION, Putative transcriptional regulator TA0346 | | Authors: | Grantz Saskova, K, Chruszcz, M, Evdokimova, E, Egorova, O, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-09-22 | | Release date: | 2008-09-30 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative transcriptional regulator TA0346 from Thermoplasma acidophilum
To be Published
|
|
3Q3C
 
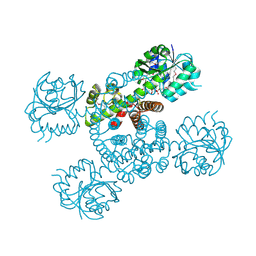 | | Crystal structure of a serine dehydrogenase from Pseudomonas aeruginosa pao1 in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Probable 3-hydroxyisobutyrate dehydrogenase | | Authors: | Tan, K, Singer, A.U, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-12-21 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Biochemical and Structural Studies of Uncharacterized Protein PA0743 from Pseudomonas aeruginosa Revealed NAD+-dependent L-Serine Dehydrogenase.
J.Biol.Chem., 287, 2012
|
|
3FYN
 
 | | Crystal structure from the mobile metagenome of Cole Harbour Salt Marsh: Integron Cassette Protein HFX_CASS3 | | Descriptor: | ACETATE ION, Integron gene cassette protein HFX_CASS3, MAGNESIUM ION | | Authors: | Sureshan, V, Deshpande, C.N, Harrop, S.J, Kudritska, M, Koenig, J.E, Evdokimova, E, Osipiuk, J, Edwards, A.M, Savchenko, A, Joachimiak, A, Doolittle, W.F, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-22 | | Release date: | 2009-02-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Structure from the mobile metagenome of Cole Harbour Salt Marsh: Integron Cassette Protein HFX_CASS3
To be Published
|
|
3EEH
 
 | |
3EFA
 
 | | Crystal structure of putative N-acetyltransferase from Lactobacillus plantarum | | Descriptor: | FORMIC ACID, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Chang, C, Li, H, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-09-08 | | Release date: | 2008-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.423 Å) | | Cite: | Crystal structure of putative N-acetyltransferase from Lactobacillus plantarum
To be Published
|
|
3N04
 
 | | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (FAMILY 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 | | Descriptor: | GLYCEROL, alpha-glucosidase | | Authors: | Tan, K, Tesar, C, Freeman, L, Wilton, R, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-13 | | Release date: | 2010-06-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (FAMILY 31) FROM RUMINOCOCCUS OBEUM ATCC 29174
Faseb J., 24, 2010
|
|
3G1J
 
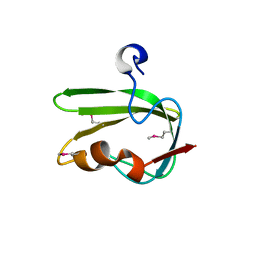 | | Structure from the mobile metagenome of Vibrio cholerae. Integron cassette protein VCH_CASS4. | | Descriptor: | Integron cassette protein | | Authors: | Deshpande, C.N, Sureshan, V, Harrop, S.J, Boucher, Y, Xu, X, Cui, H, Edwards, A, Savchenko, A, Joachimiak, A, Tan, K, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-30 | | Release date: | 2009-02-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure from the mobile metagenome of Vibrio cholerae. Integron cassette protein VCH_CASS4.
To be published
|
|
3NA2
 
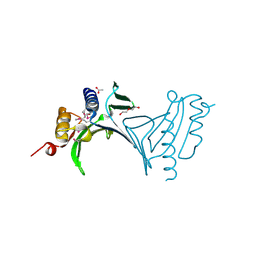 | | Crystal Structure of Protein of Unknown Function from Mine Drainage Metagenome Leptospirillum rubarum | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, Uncharacterized protein | | Authors: | Kim, Y, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-31 | | Release date: | 2010-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Crsystal Structure of Protein of Unknown Function from Mine Drainage Metagenome Leptospirillum rubarum
To be Published
|
|
3G64
 
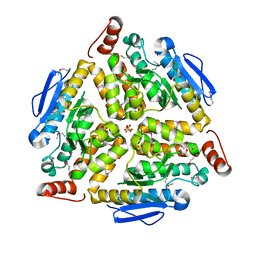 | | Crystal structure of putative enoyl-CoA hydratase from Streptomyces coelicolor A3(2) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Putative enoyl-CoA hydratase, ... | | Authors: | Kim, Y, Xu, X, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-06 | | Release date: | 2009-03-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of Putative Enoyl-CoA Hydratase from Streptomyces coelicolor A3(2)
To be Published
|
|
3EUP
 
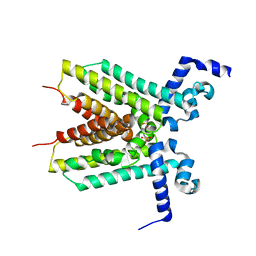 | | The crystal structure of the transcriptional regulator, TetR family from Cytophaga hutchinsonii | | Descriptor: | SULFATE ION, Transcriptional regulator, TetR family | | Authors: | Zhang, R, Bigelow, L, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-10 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of the transcriptional regulator, TetR family from Cytophaga hutchinsonii
To be Published
|
|
3NE7
 
 | | Crystal structure of paia n-acetyltransferase from thermoplasma acidophilum in complex with coenzyme a | | Descriptor: | ACETYLTRANSFERASE, BETA-MERCAPTOETHANOL, COENZYME A, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, F.W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-08 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the novel PaiA N-acetyltransferase from Thermoplasma acidophilum involved in the negative control of sporulation and degradative enzyme production.
Proteins, 79, 2011
|
|
4LMI
 
 | |
