4JU6
 
 | | Crystal structure of hcv ns5b polymerase in complex with compound 24 | | Descriptor: | 2-{[(trans-4-methylcyclohexyl)carbonyl](propan-2-yl)amino}-5-phenoxybenzoic acid, Genome polyprotein, MAGNESIUM ION | | Authors: | Coulombe, R. | | Deposit date: | 2013-03-24 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anthranilic acid-based Thumb Pocket 2 HCV NS5B polymerase inhibitors with sub-micromolar potency in the cell-based replicon assay.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2FE3
 
 | |
3RH0
 
 | |
4JU7
 
 | | Crystal structure of hcv ns5b polymerase in complex with compound 24 | | Descriptor: | 2-{[(trans-4-methylcyclohexyl)carbonyl](propan-2-yl)amino}-5-phenoxybenzoic acid, Genome polyprotein, MAGNESIUM ION | | Authors: | Coulombe, R. | | Deposit date: | 2013-03-24 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anthranilic acid-based Thumb Pocket 2 HCV NS5B polymerase inhibitors with sub-micromolar potency in the cell-based replicon assay.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3QOQ
 
 | |
5OP7
 
 | | Structure of CHK1 10-pt. mutant complex with pyrrolopyrimidine LRRK2 inhibitor | | Descriptor: | CHLORIDE ION, SODIUM ION, Serine/threonine-protein kinase Chk1, ... | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-09 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
5LAC
 
 | |
4JU1
 
 | | Crystal structure of hcv ns5b polymerase in complex with compound 6 | | Descriptor: | 6-[3-([1,3]oxazolo[4,5-b]pyridin-2-yl)-2-(trifluoromethyl)phenoxy]-1-(2,4,6-trifluorobenzyl)quinazolin-4(1H)-one, GLYCEROL, Genome polyprotein, ... | | Authors: | Coulombe, R. | | Deposit date: | 2013-03-24 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Dynamics Simulations and Structure-Based Rational Design Lead to Allosteric HCV NS5B Polymerase Thumb Pocket 2 Inhibitor with Picomolar Cellular Replicon Potency.
J.Med.Chem., 57, 2014
|
|
5OOP
 
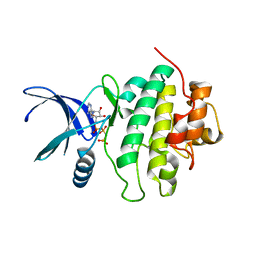 | | Structure of CHK1 10-pt. mutant complex with AMP-PNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-08 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
5OP5
 
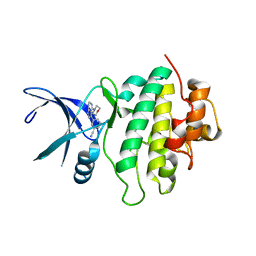 | | Structure of CHK1 10-pt. mutant complex with pyrrolopyrimidine LRRK2 inhibitor | | Descriptor: | 3-[4-(morpholin-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-09 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
5LAK
 
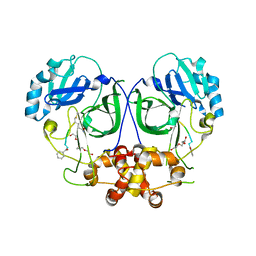 | | Ligand-bound structure of Cavally Virus 3CL Protease | | Descriptor: | 3Cl Protease, BEZ-TYR-TYR-ASN-ECC Peptide inhibitor, PENTAETHYLENE GLYCOL, ... | | Authors: | Kanitz, M, Heine, A, Diederich, W.E. | | Deposit date: | 2016-06-14 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural basis for catalysis and substrate specificity of a 3C-like cysteine protease from a mosquito mesonivirus.
Virology, 533, 2019
|
|
5OPR
 
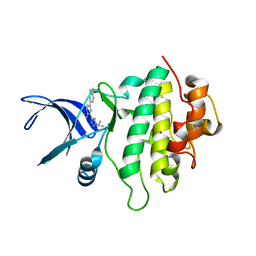 | | Structure of CHK1 10-pt. mutant complex with aminopyridine LRRK2 inhibitor | | Descriptor: | 5-[4-(morpholin-4-ylmethyl)phenyl]-3-(1-propan-2-yl-1,2,3-triazol-4-yl)pyridin-2-amine, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-10 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
5OPV
 
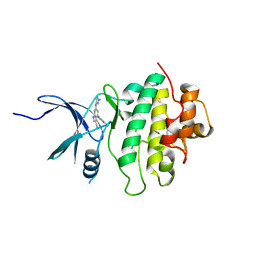 | | Structure of CHK1 10-pt. mutant complex with pyrrolopyridine LRRK2 inhibitor | | Descriptor: | 4-(3-methylphenyl)-6-[(1-methylpyrazol-3-yl)amino]-1~{H}-pyrrolo[2,3-b]pyridine-3-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-10 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
5OQ5
 
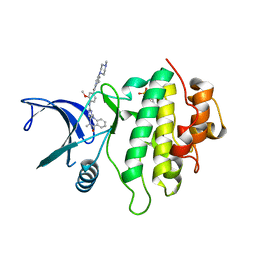 | | Structure of CHK1 8-pt. mutant complex with aminopyrimido-benzodiazepinone LRRK2 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[(2-methoxy-4-{[4-(4-methylpiperazin-1-yl)piperidin-1-yl]carbonyl}phenyl)amino]-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-10 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
1T6P
 
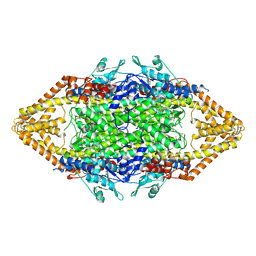 | |
5MPV
 
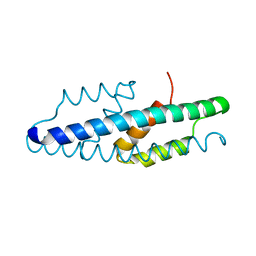 | |
1T6J
 
 | |
1ZBN
 
 | |
1DYR
 
 | | THE STRUCTURE OF PNEUMOCYSTIS CARINII DIHYDROFOLATE REDUCTASE TO 1.9 ANGSTROMS RESOLUTION | | Descriptor: | DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRIMETHOPRIM | | Authors: | Champness, J.N, Achari, A, Ballantine, S.P, Bryant, P.K, Delves, C.J, Stammers, D.K. | | Deposit date: | 1994-09-14 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The structure of Pneumocystis carinii dihydrofolate reductase to 1.9 A resolution.
Structure, 2, 1994
|
|
5LV0
 
 | | Structure of Human Neurolysin (E475Q) in complex with amyloid-beta 35-40 peptide product | | Descriptor: | CHLORIDE ION, GLY-VAL-VAL amyloid 35-40 fragment, Neurolysin, ... | | Authors: | Masuyer, G, Berntsson, R.P.-A, Teixeira, P.F, Kmiec, B, Glaser, E, Stenmark, P. | | Deposit date: | 2016-09-12 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional analysis of Neurolysin, a new component of the mitochondrial peptidolytic network
To Be Published
|
|
4JTZ
 
 | | Crystal structure of hcv ns5b polymerase in complex with compound 4 | | Descriptor: | 3-{[4-oxo-1-(2,4,6-trifluorobenzyl)-1,4-dihydroquinazolin-6-yl]oxy}-N-(pyridin-3-yl)-2-(trifluoromethyl)benzamide, GLYCEROL, Genome polyprotein, ... | | Authors: | Coulombe, R. | | Deposit date: | 2013-03-24 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular Dynamics Simulations and Structure-Based Rational Design Lead to Allosteric HCV NS5B Polymerase Thumb Pocket 2 Inhibitor with Picomolar Cellular Replicon Potency.
J.Med.Chem., 57, 2014
|
|
3C22
 
 | |
5OP4
 
 | | Structure of CHK1 10-pt. mutant complex with aminopyrimidine LRRK2 inhibitor | | Descriptor: | Serine/threonine-protein kinase Chk1, [4-[[4-(ethylamino)-5-(trifluoromethyl)pyrimidin-2-yl]amino]-2-fluoranyl-5-methoxy-phenyl]-morpholin-4-yl-methanone | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-09 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
5OPS
 
 | | Structure of CHK1 10-pt. mutant complex with pyrrolopyridine LRRK2 inhibitor | | Descriptor: | 4-(3-hydroxyphenyl)-1~{H}-pyrrolo[2,3-b]pyridine-3-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-10 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
5OOR
 
 | | Structure of CHK1 10-pt. mutant complex with staurosporine | | Descriptor: | CHLORIDE ION, STAUROSPORINE, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-08 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
