6YOQ
 
 | |
6YNJ
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_046 | | Descriptor: | (2~{R})-2-(2-chlorophenyl)-5,5-dimethyl-morpholine, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YNM
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_096 | | Descriptor: | 1-[(2~{R},5~{S})-2-(3-chlorophenyl)-5-methyl-morpholin-4-yl]-3-methoxy-propan-2-ol, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YKI
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_092 | | Descriptor: | SULFATE ION, YTHDC1, ~{N}-ethyl-2-[(2~{S},5~{R})-5-methyl-2-phenyl-morpholin-4-yl]ethanamine | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YKJ
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_125 | | Descriptor: | SULFATE ION, YTHDC1, ~{N}-methyl-4-(methylamino)pyridine-2-carboxamide | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YNI
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_036 | | Descriptor: | (2~{R},3~{R})-2-(3-chlorophenyl)-3,5,5-trimethyl-morpholine, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YNL
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_078 | | Descriptor: | 3-[(2~{R})-5,5-dimethyl-2-phenyl-morpholin-4-yl]-~{N}-(methylcarbamoyl)propanamide, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YNP
 
 | | Crystal structure of YTHDC1 with compound T96C1 | | Descriptor: | CHLORIDE ION, SULFATE ION, YTHDC1, ... | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YL8
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_011 | | Descriptor: | (2~{R},5~{S})-2-(2-chlorophenyl)-5-methyl-morpholine, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YNO
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_139 | | Descriptor: | 6-[[methyl-[(1-phenylpyrazol-3-yl)methyl]amino]methyl]-1~{H}-pyrimidine-2,4-dione, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
6ZD9
 
 | | Crystal structure of YTHDC1 apo purified using GST tag | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Caflisch, A. | | Deposit date: | 2020-06-14 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
8WNS
 
 | | Cryo EM map of SLC7A10 in the apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Amino acid transporter heavy chain SLC3A2, Asc-type amino acid transporter 1 | | Authors: | Li, Y.N, Guo, Y.Y, Dai, L, Yan, R.H. | | Deposit date: | 2023-10-06 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Cryo-EM structure of the human Asc-1 transporter complex.
Nat Commun, 15, 2024
|
|
8WNY
 
 | | Cryo EM map of SLC7A10-SLC3A2 complex in the D-serine bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Amino acid transporter heavy chain SLC3A2, Asc-type amino acid transporter 1, ... | | Authors: | Li, Y.N, Guo, Y.Y, Dai, L, Yan, R.H. | | Deposit date: | 2023-10-06 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the human Asc-1 transporter complex.
Nat Commun, 15, 2024
|
|
8WNT
 
 | | Cryo EM map of SLC7A10 with L-Alanine substrate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALANINE, ... | | Authors: | Li, Y.N, Guo, Y.Y, Dai, L, Yan, R.H. | | Deposit date: | 2023-10-06 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Cryo-EM structure of the human Asc-1 transporter complex.
Nat Commun, 15, 2024
|
|
4HRO
 
 | |
4HRS
 
 | |
3K8S
 
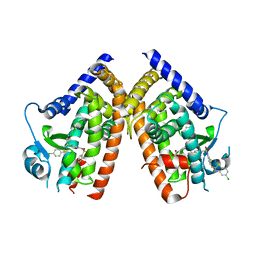 | | Crystal Structure of PPARg in complex with T2384 | | Descriptor: | 2-chloro-N-{3-chloro-4-[(5-chloro-1,3-benzothiazol-2-yl)sulfanyl]phenyl}-4-(trifluoromethyl)benzenesulfonamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Wang, Z. | | Deposit date: | 2009-10-14 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | T2384, a novel antidiabetic agent with unique peroxisome proliferator-activated receptor gamma binding properties
J.Biol.Chem., 283, 2008
|
|
8I91
 
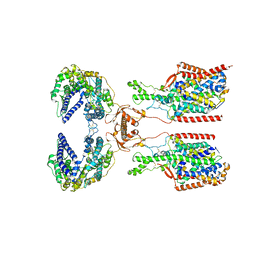 | | ACE2-SIT1 complex bound with proline | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Zhang, Y.Y, Shen, Y.P, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insight into the substrate recognition and transport mechanism of amino acid transporter complex ACE2-B 0 AT1 and ACE2-SIT1.
Cell Discov, 9, 2023
|
|
8I93
 
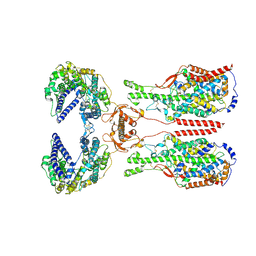 | | ACE2-B0AT1 complex bound with methionine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Zhang, Y.Y, Shen, Y.P, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into the substrate recognition and transport mechanism of amino acid transporter complex ACE2-B 0 AT1 and ACE2-SIT1.
Cell Discov, 9, 2023
|
|
8I92
 
 | | ACE2-B0AT1 complex bound with glutamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Zhang, Y.Y, Shen, Y.P, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insight into the substrate recognition and transport mechanism of amino acid transporter complex ACE2-B 0 AT1 and ACE2-SIT1.
Cell Discov, 9, 2023
|
|
4F55
 
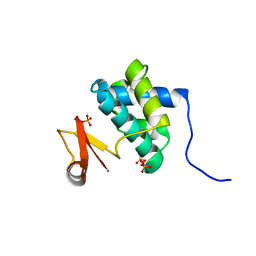 | | Crystal Structure of the Catalytic Domain of the Bacillus cereus SleB Protein | | Descriptor: | PHOSPHATE ION, Spore cortex-lytic enzyme | | Authors: | Hao, B. | | Deposit date: | 2012-05-11 | | Release date: | 2012-07-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Bacillus cereus SleB Protein, Important in Cortex Peptidoglycan Degradation during Spore Germination.
J.Bacteriol., 194, 2012
|
|
6UUQ
 
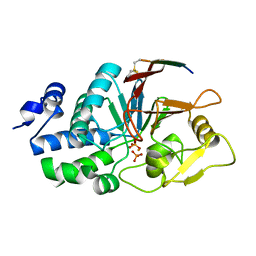 | | Structure of Calcineurin bound to RCAN1 | | Descriptor: | Calcipressin-1, FE (III) ION, PHOSPHATE ION, ... | | Authors: | Sheftic, S, Page, R, Peti, W. | | Deposit date: | 2019-10-31 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | The structure of the RCAN1:CN complex explains the inhibition of and substrate recruitment by calcineurin.
Sci Adv, 6, 2020
|
|
3V72
 
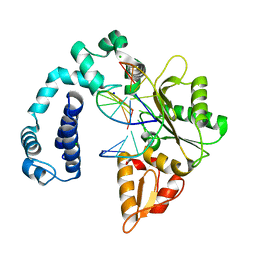 | | Crystal Structure of Rat DNA polymerase beta Mutator E295K: Enzyme-dsDNA | | Descriptor: | CHLORIDE ION, DNA 5'-D(P*AP*AP*AP*CP*TP*CP*AP*CP*AP*T)-3', DNA 5'-D(P*AP*TP*GP*TP*GP*AP*GP*T)-3', ... | | Authors: | Gridley, C.L, Jaeger, J. | | Deposit date: | 2011-12-20 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Unfavorable Electrostatic and Steric Interactions in DNA Polymerase beta E295K Mutant Interfere with the Enzyme s Pathway
J.Am.Chem.Soc., 134, 2012
|
|
6LYP
 
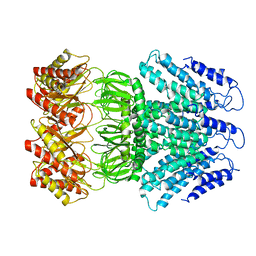 | | Cryo-EM structure of AtMSL1 wild type | | Descriptor: | Mechanosensitive ion channel protein 1, mitochondrial | | Authors: | Sun, L. | | Deposit date: | 2020-02-15 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural Insights into a Plant Mechanosensitive Ion Channel MSL1.
Cell Rep, 30, 2020
|
|
8FR8
 
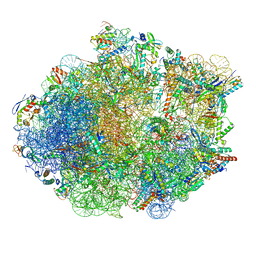 | | Structure of Mycobacterium smegmatis Rsh bound to a 70S translation initiation complex | | Descriptor: | 16S rRNA (1511-MER), 23S rRNA (3119-MER), 30S ribosomal protein S10, ... | | Authors: | Majumdar, S, Sharma, M.R, Manjari, S.R, Banavali, N.K, Agrawal, R.K. | | Deposit date: | 2023-01-06 | | Release date: | 2023-05-17 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Starvation sensing by mycobacterial RelA/SpoT homologue through constitutive surveillance of translation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
