1Z6B
 
 | | Crystal structure of Plasmodium falciparum FabZ at 2.1 A | | Descriptor: | CACODYLATE ION, CHLORIDE ION, SULFATE ION, ... | | Authors: | Kostrewa, D, Winkler, F.K, Folkers, G, Scapozza, L, Perozzo, R. | | Deposit date: | 2005-03-22 | | Release date: | 2005-06-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The crystal structure of PfFabZ, the unique beta-hydroxyacyl-ACP dehydratase involved in fatty acid biosynthesis of Plasmodium falciparum
PROTEIN SCI., 14, 2005
|
|
6S0Z
 
 | | Erythromycin Resistant Staphylococcus aureus 50S ribosome (delta R88 A89 uL22) in complex with erythromycin. | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Halfon, Y, Matozv, D, Eyal, Z, Bashan, A, Zimmerman, E, Kjeldgaard, J, Ingmer, H, Yonath, A. | | Deposit date: | 2019-06-18 | | Release date: | 2019-08-21 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Exit tunnel modulation as resistance mechanism of S. aureus erythromycin resistant mutant.
Sci Rep, 9, 2019
|
|
6WYC
 
 | |
6WZ6
 
 | | Complex of mutant (K173M) of Pseudomonas 7A Glutaminase-Asparaginase with L-Glu at pH 5. Covalent acyl-enzyme intermediate | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, Glutaminase-asparaginase | | Authors: | Strzelczyk, P, Zhang, D, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2020-05-13 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Generalized enzymatic mechanism of catalysis by tetrameric L-asparaginases from mesophilic bacteria.
Sci Rep, 10, 2020
|
|
1Z58
 
 | | Crystal structure of a complex of the ribosome large subunit with rapamycin | | Descriptor: | 23S RIBOSOMAL RNA, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Amit, M, Berisio, R, Baram, D, Harms, J, Bashan, A, Yonath, A. | | Deposit date: | 2005-03-17 | | Release date: | 2005-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | A crevice adjoining the ribosome tunnel: Hints for cotranslational folding.
Febs Lett., 579, 2005
|
|
3ZZK
 
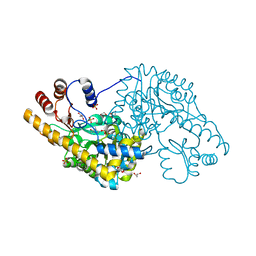 | | Structure of an engineered aspartate aminotransferase | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE, GLYCEROL, ... | | Authors: | Fernandez, F.J, deVries, D, Pena-Soler, E, Coll, M, Christen, P, Gehring, H, Vega, M.C. | | Deposit date: | 2011-09-01 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure and Mechanism of a Cysteine Sulfinate Desulfinase Engineered on the Aspartate Aminotransferase Scaffold.
Biocim.Biophys.Acta, 1824, 2011
|
|
1ZEY
 
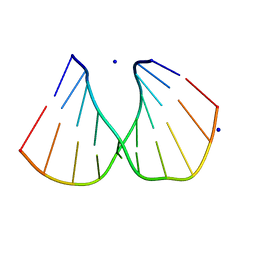 | | CGG A-DNA | | Descriptor: | 5'-D(*CP*CP*CP*CP*GP*CP*GP*GP*GP*G)-3', SODIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-19 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZF6
 
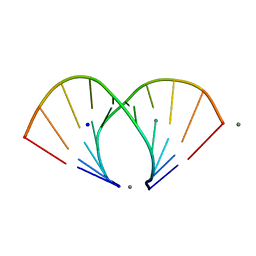 | | TGG DUPLEX A-DNA | | Descriptor: | 5'-D(*CP*CP*CP*CP*AP*TP*GP*GP*GP*G)-3', CALCIUM ION, SODIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-19 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
7UV5
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S/D286N mutant, in complex with a Lys48-linked di-ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, Papain-like protease nsp3, Ubiquitin, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lanham, B.T, Wydorski, P, Fushman, D, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-04-29 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
1ZFJ
 
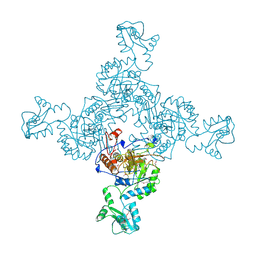 | | INOSINE MONOPHOSPHATE DEHYDROGENASE (IMPDH; EC 1.1.1.205) FROM STREPTOCOCCUS PYOGENES | | Descriptor: | INOSINE MONOPHOSPHATE DEHYDROGENASE, INOSINIC ACID | | Authors: | Zhang, R, Evans, G, Rotella, F.J, Westbrook, E.M, Beno, D, Huberman, E, Joachimiak, A, Collart, F.R. | | Deposit date: | 1999-03-29 | | Release date: | 2000-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characteristics and crystal structure of bacterial inosine-5'-monophosphate dehydrogenase.
Biochemistry, 38, 1999
|
|
4RQN
 
 | | Crystal structure of the native BICC1 SAM Domain R924E mutant | | Descriptor: | Protein bicaudal C homolog 1, ZINC ION | | Authors: | Leettola, C.N, Cascio, D, Bowie, J.U. | | Deposit date: | 2014-11-03 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Bicc1 SAM Polymer and Mapping of Interactions between the Ciliopathy-Associated Proteins Bicc1, ANKS3, and ANKS6.
Structure, 26, 2018
|
|
1ZKF
 
 | |
6TPW
 
 | | Crystal structures of FNIII domain one through four of the human leucocyte common antigen-related protein ( LAR) | | Descriptor: | Receptor-type tyrosine-protein phosphatase F, SULFATE ION | | Authors: | Vilstrup, J.P, Thirup, S.S, Simonsen, A, Birkefeldt, T, Strandbygaard, D. | | Deposit date: | 2019-12-14 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal and solution structures of fragments of the human leucocyte common antigen-related protein.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6VOJ
 
 | | Chloroplast ATP synthase (R3, CF1FO) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-30 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.34 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
1ZOY
 
 | | Crystal Structure of Mitochondrial Respiratory Complex II from porcine heart at 2.4 Angstroms | | Descriptor: | FAD-binding protein, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Sun, F, Huo, X, Zhai, Y, Wang, A, Xu, J, Su, D, Bartlam, M, Rao, Z. | | Deposit date: | 2005-05-15 | | Release date: | 2005-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Mitochondrial Respiratory Membrane Protein Complex II
Cell(Cambridge,Mass.), 121, 2005
|
|
6VTH
 
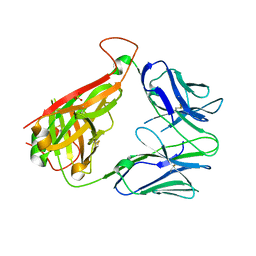 | | p53-specific T cell receptor | | Descriptor: | SULFATE ION, T-cell Receptor 12-6, Alfa chain, ... | | Authors: | Wu, D, Gallagher, D.T, Gowthaman, R, Pierce, B.G, Mariuzza, R.A. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural basis for oligoclonal T cell recognition of a shared p53 cancer neoantigen.
Nat Commun, 11, 2020
|
|
4Q3G
 
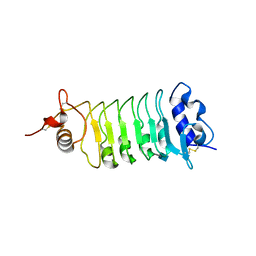 | | Structure of the OsSERK2 leucine rich repeat extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, OsSERK2 | | Authors: | McAndrew, R.P, Pruitt, R.N, Kamita, S.G, Pereira, J.H, Majumder, D, Hammock, B.D, Adams, P.D, Ronald, P.C. | | Deposit date: | 2014-04-11 | | Release date: | 2014-11-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.787 Å) | | Cite: | Structure of the OsSERK2 leucine-rich repeat extracellular domain.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4QHL
 
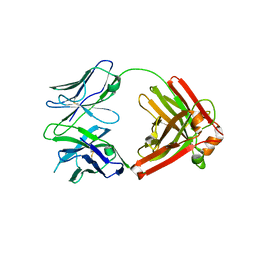 | | I3.2 (unbound) from CH103 Lineage | | Descriptor: | I3 heavy chain, UCA light chain | | Authors: | Fera, D, Harrison, S.C. | | Deposit date: | 2014-05-28 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.153 Å) | | Cite: | Affinity maturation in an HIV broadly neutralizing B-cell lineage through reorientation of variable domains.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1Z19
 
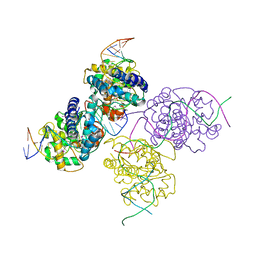 | | Crystal structure of a lambda integrase(75-356) dimer bound to a COC' core site | | Descriptor: | 33-MER, 5'-D(*CP*TP*CP*GP*TP*TP*CP*AP*GP*CP*TP*TP*TP*TP*TP*T)-3', 5'-D(P*TP*TP*TP*AP*TP*AP*CP*TP*AP*AP*GP*TP*TP*GP*GP*CP*AP*TP*TP*A)-3', ... | | Authors: | Biswas, T, Aihara, H, Radman-Livaja, M, Filman, D, Landy, A, Ellenberger, T. | | Deposit date: | 2005-03-03 | | Release date: | 2005-06-28 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A structural basis for allosteric control of DNA recombination by lambda integrase.
Nature, 435, 2005
|
|
6VOM
 
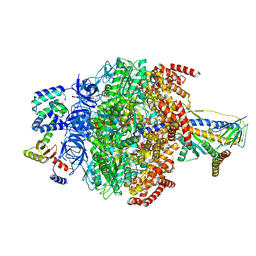 | | Chloroplast ATP synthase (R2, CF1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-30 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
1XV2
 
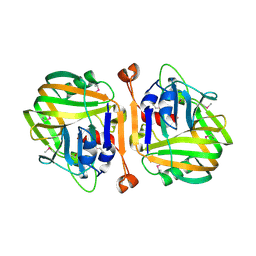 | | Crystal Structure of a Protein of Unknown Function Similar to Alpha-acetolactate Decarboxylase from Staphylococcus aureus | | Descriptor: | ZINC ION, hypothetical protein, similar to alpha-acetolactate decarboxylase | | Authors: | Duke, N.E.C, Zhang, R, Quartey, P, Collart, F, Holzle, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-10-26 | | Release date: | 2004-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a hypothetical protein similar to alpha-acetolactate
To be Published
|
|
6VTU
 
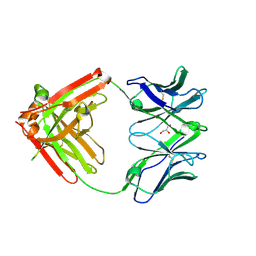 | | DH717.1 Fab monomer in complex with man9 glycan | | Descriptor: | DH717.1 heavy chain, DH717.1 light chain, GLYCEROL, ... | | Authors: | Fera, D, Bronkema, N. | | Deposit date: | 2020-02-13 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
1XWE
 
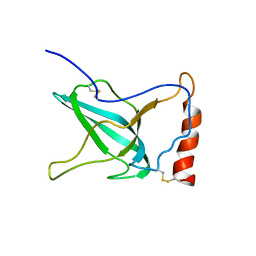 | | NMR Structure of C345C (NTR) domain of C5 of complement | | Descriptor: | Complement C5 | | Authors: | Bramham, J, Thai, C.-T, Soares, D.C, Uhrin, D, Ogata, R.T, Barlow, P.N. | | Deposit date: | 2004-10-30 | | Release date: | 2004-12-21 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Functional Insights from the Structure of the Multifunctional C345C Domain of C5 of Complement
J.Biol.Chem., 280, 2005
|
|
4QNR
 
 | | CRYSTAL STRUCTURE OF PSPF(1-265) E108Q MUTANT bound to ATP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Darbari, V.C, Lawton, E, Lu, D, Burrows, P.C, Wiesler, S, Joly, N, Zhang, N, Zhang, X, Buck, M. | | Deposit date: | 2014-06-18 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | Molecular basis of nucleotide-dependent substrate engagement and remodeling by an AAA+ activator.
Nucleic Acids Res., 42, 2014
|
|
1XYY
 
 | | Low Temperature (100K) Crystal Structure Of Flavodoxin Mutant S64C, homodimer, oxidised state | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Artali, R, Marchini, N, Meneghetti, F, Cavazzini, D, Bombieri, G, Rossi, G.L, Gilardi, G. | | Deposit date: | 2004-11-11 | | Release date: | 2004-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Redox properties and crystal structures of a Desulfovibrio vulgaris flavodoxin mutant in the monomeric and homodimeric forms.
Biochim.Biophys.Acta, 1794, 2009
|
|
