4X9D
 
 | | High-resolution structure of Hfq from Methanococcus jannaschii in complex with UMP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nikulin, A.D, Tishchenko, S.V, Nikonova, E.Y, Murina, V.N, Mihailina, A.O, Lekontseva, N.V. | | Deposit date: | 2014-12-11 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization of RNA-binding properties of the archaeal Hfq-like protein from Methanococcus jannaschii.
J. Biomol. Struct. Dyn., 35, 2017
|
|
1XS0
 
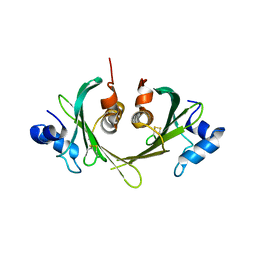 | | Structure of the E. coli Ivy protein | | Descriptor: | Inhibitor of vertebrate lysozyme | | Authors: | Abergel, C, Monchois, V, Byrn, D, Lazzaroni, J.C, Claverie, J.M. | | Deposit date: | 2004-10-18 | | Release date: | 2004-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure and evolution of the Ivy protein family, unexpected lysozyme inhibitors in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
3NN0
 
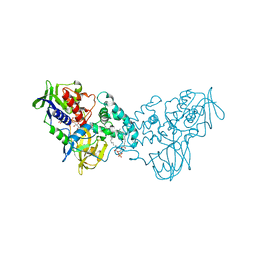 | | Complex of 6-hydroxy-L-nicotine oxidase with nicotinamide | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 6-hydroxy-L-nicotine oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2010-06-23 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure analysis of free and substrate-bound 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
3NK2
 
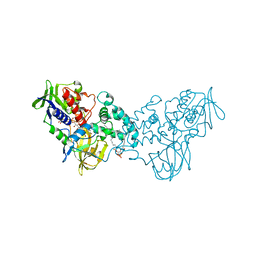 | | Complex of 6-hydroxy-L-nicotine oxidase with dopamine | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 6-hydroxy-L-nicotine oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2010-06-18 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure analysis of free and substrate-bound 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
3NGC
 
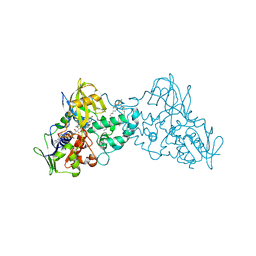 | | Complex of 6-hydroxy-L-nicotine oxidase with intermediate methylmyosmine product formed during catalytic turnover | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 5-(1-methyl-4,5-dihydro-1H-pyrrol-2-yl)pyridin-2-ol, 5-[(2S)-1-methylpyrrolidin-2-yl]pyridin-2-ol, ... | | Authors: | Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2010-06-11 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure analysis of free and substrate-bound 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
3NH3
 
 | |
3NHO
 
 | | Complex of 6-hydroxy-L-nicotine oxidase with product bound at active site | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 1-(6-hydroxypyridin-3-yl)-4-(methylamino)butan-1-one, 6-hydroxy-L-nicotine oxidase, ... | | Authors: | Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2010-06-14 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure analysis of free and substrate-bound 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
3NK1
 
 | | Complex of 6-hydroxy-L-nicotine oxidase with serotonin | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 6-hydroxy-L-nicotine oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2010-06-18 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure analysis of free and substrate-bound 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
3NN6
 
 | | Crystal structure of inhibitor-bound in active centre 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, (2R,3S,4S)-5-({[(acetylcarbamoyl)amino]methyl}[(3S,4R)-6-amino-3,4-dimethylhexyl]amino)-2,3,4-trihydroxypentyl [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name), 5-[(2R)-1-methylpyrrolidin-2-yl]pyridin-2-ol, ... | | Authors: | Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2010-06-23 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure analysis of free and substrate-bound 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
3NK0
 
 | |
5LHL
 
 | |
6RHQ
 
 | | Crystal Structure of Two-Domain Laccase mutant I170A from Streptomyces griseoflavus | | Descriptor: | COPPER (II) ION, GLYCEROL, SULFATE ION, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Kolyadenko, I.A. | | Deposit date: | 2019-04-22 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Investigations of Accessibility of T2/T3 Copper Center of Two-Domain Laccase fromStreptomyces griseoflavusAc-993.
Int J Mol Sci, 20, 2019
|
|
7XQV
 
 | | The complex of nanobody Rh57 binding to GTP-bound RhoA active form | | Descriptor: | ALANINE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Zhang, Y.R, Liu, R, Ding, Y. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural insights into the binding of nanobody Rh57 to active RhoA-GTP.
Biochem.Biophys.Res.Commun., 616, 2022
|
|
6S0O
 
 | | Crystal Structure of Two-Domain Laccase from Streptomyces griseoflavus produced at 0.25 mM copper sulfate in growth medium | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Kolyadenko, I.A. | | Deposit date: | 2019-06-17 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Investigations of Accessibility of T2/T3 Copper Center of Two-Domain Laccase fromStreptomyces griseoflavusAc-993.
Int J Mol Sci, 20, 2019
|
|
6RH9
 
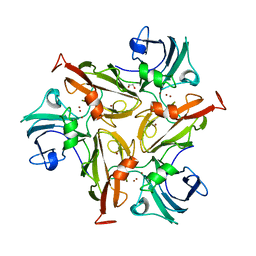 | | Crystal Structure of Two-Domain Laccase mutant I170F from Streptomyces griseoflavus | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Kolyadenko, I.A. | | Deposit date: | 2019-04-19 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Investigations of Accessibility of T2/T3 Copper Center of Two-Domain Laccase fromStreptomyces griseoflavusAc-993.
Int J Mol Sci, 20, 2019
|
|
6YC5
 
 | |
6YBI
 
 | |
6YBO
 
 | |
6YBX
 
 | |
6K2U
 
 | | Crystal structure of Thr66 ADP-ribosylated ubiquitin | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, MAGNESIUM ION, Polyubiquitin-C, ... | | Authors: | Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2019-05-15 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | Threonine ADP-Ribosylation of Ubiquitin by a Bacterial Effector Family Blocks Host Ubiquitination.
Mol.Cell, 78, 2020
|
|
6IHJ
 
 | |
6YBR
 
 | |
6YBF
 
 | |
5M41
 
 | | Crystal structure of nigritoxine | | Descriptor: | MAGNESIUM ION, Nigritoxine | | Authors: | Czjzek, M, Labreuche, L, Jeudy, A, Le Roux, F. | | Deposit date: | 2016-10-17 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nigritoxin is a bacterial toxin for crustaceans and insects.
Nat Commun, 8, 2017
|
|
1EG0
 
 | | FITTING OF COMPONENTS WITH KNOWN STRUCTURE INTO AN 11.5 A CRYO-EM MAP OF THE E.COLI 70S RIBOSOME | | Descriptor: | FORMYL-METHIONYL-TRNA, FRAGMENT OF 16S RRNA HELIX 23, FRAGMENT OF 23S RRNA, ... | | Authors: | Gabashvili, I.S, Agrawal, R.K, Spahn, C.M.T, Grassucci, R.A, Svergun, D.I, Frank, J, Penczek, P. | | Deposit date: | 2000-02-11 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Solution structure of the E. coli 70S ribosome at 11.5 A resolution.
Cell(Cambridge,Mass.), 100, 2000
|
|
