7YQE
 
 | |
8YAG
 
 | | Cryo-electron microscopic structure of an amide hydrolase from Pseudoxanthomonas wuyuanensis | | Descriptor: | Imidazolonepropionase, ZINC ION | | Authors: | Dai, L.H, Xu, Y.H, Hu, Y.M, Niu, D, Yang, X.C, Shen, P.P, Li, X, Xie, Z.Z, Li, H, Guo, R.-T, Chen, C.-C. | | Deposit date: | 2024-02-09 | | Release date: | 2024-10-09 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Functional characterization and structural basis of an efficient ochratoxin A-degrading amidohydrolase.
Int.J.Biol.Macromol., 278, 2024
|
|
7XFG
 
 | |
6DBM
 
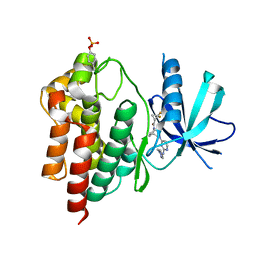 | | Tyk2 with compound 23 | | Descriptor: | Non-receptor tyrosine-protein kinase TYK2, [(1S)-2,2-difluorocyclopropyl][(1R,5S)-3-{2-[(1-methyl-1H-pyrazol-4-yl)amino]pyrimidin-4-yl}-3,8-diazabicyclo[3.2.1]octan-8-yl]methanone | | Authors: | Vajdos, F.F. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.368 Å) | | Cite: | Dual Inhibition of TYK2 and JAK1 for the Treatment of Autoimmune Diseases: Discovery of (( S)-2,2-Difluorocyclopropyl)((1 R,5 S)-3-(2-((1-methyl-1 H-pyrazol-4-yl)amino)pyrimidin-4-yl)-3,8-diazabicyclo[3.2.1]octan-8-yl)methanone (PF-06700841).
J. Med. Chem., 61, 2018
|
|
8YAK
 
 | | Cryo-EM structure and rational engineering of a novel efficient ochratoxin A-detoxifying amidohydrolase | | Descriptor: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, Imidazolonepropionase, ZINC ION | | Authors: | Dai, L.H, Xu, Y.H, Hu, Y.M, Niu, D, Yang, X.C, Shen, P.P, Li, X, Xie, Z.Z, Li, H, Guo, R.-T, Chen, C.-C. | | Deposit date: | 2024-02-09 | | Release date: | 2024-12-18 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | Functional characterization and structural basis of an efficient ochratoxin A-degrading amidohydrolase.
Int.J.Biol.Macromol., 278, 2024
|
|
7KL8
 
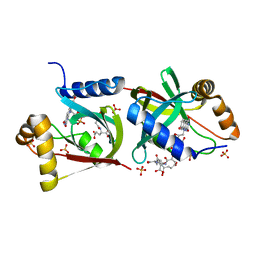 | | Structure of F420 binding protein Rv1558 from Mycobacterium tuberculosis with F420 bound | | Descriptor: | COENZYME F420, COENZYME F420-3, Deazaflavin-dependent nitroreductase, ... | | Authors: | Lee, B.M, Tan, L.L, Jackson, C.J. | | Deposit date: | 2020-10-29 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.469 Å) | | Cite: | Potency boost of a Mycobacterium tuberculosis dihydrofolate reductase inhibitor by multienzyme F 420 H 2 -dependent reduction.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6DBN
 
 | | Jak1 with compound 23 | | Descriptor: | Tyrosine-protein kinase JAK1, [(1S)-2,2-difluorocyclopropyl][(1R,5S)-3-{2-[(1-methyl-1H-pyrazol-4-yl)amino]pyrimidin-4-yl}-3,8-diazabicyclo[3.2.1]octan-8-yl]methanone | | Authors: | Vajdos, F.F. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Dual Inhibition of TYK2 and JAK1 for the Treatment of Autoimmune Diseases: Discovery of (( S)-2,2-Difluorocyclopropyl)((1 R,5 S)-3-(2-((1-methyl-1 H-pyrazol-4-yl)amino)pyrimidin-4-yl)-3,8-diazabicyclo[3.2.1]octan-8-yl)methanone (PF-06700841).
J. Med. Chem., 61, 2018
|
|
9JAP
 
 | | Helical structure of EfAvs5(SIR2-STAND) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SIR2-like domain-containing protein | | Authors: | Wang, Y, Zheng, J. | | Deposit date: | 2024-08-25 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Filamentation activates bacterial Avs5 antiviral protein.
Nat Commun, 16, 2025
|
|
7MP3
 
 | |
9ISN
 
 | |
8FJB
 
 | |
8FJA
 
 | |
6IO6
 
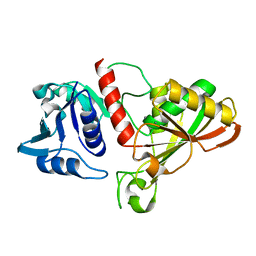 | |
5Y2F
 
 | | Human SIRT6 in complex with allosteric activator MDL-801 | | Descriptor: | 5-[[3,5-bis(chloranyl)phenyl]sulfonylamino]-2-[(5-bromanyl-4-fluoranyl-2-methyl-phenyl)sulfamoyl]benzoic acid, 9-mer peptide QTARKSTGG, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, J, Huang, Z, Song, K. | | Deposit date: | 2017-07-25 | | Release date: | 2018-11-07 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Identification of a cellularly active SIRT6 allosteric activator.
Nat. Chem. Biol., 14, 2018
|
|
6J72
 
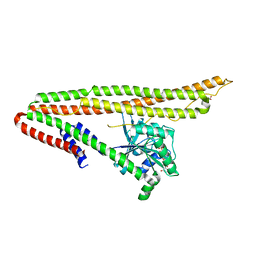 | | Crystal structure of IniA from Mycobacterium smegmatis with GTP bound | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Isoniazid inducible gene protein IniA, L(+)-TARTARIC ACID, ... | | Authors: | Wang, M.F, Guo, X.Y, Hu, J.J, Li, J, Rao, Z.H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mycobacterial dynamin-like protein IniA mediates membrane fission.
Nat Commun, 10, 2019
|
|
2IWQ
 
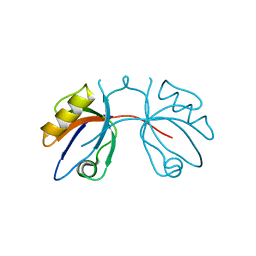 | | 7th PDZ domain of Multiple PDZ Domain Protein MPDZ | | Descriptor: | MULTIPLE PDZ DOMAIN PROTEIN | | Authors: | Elkins, J.M, Berridge, G, Savitsky, P, Smee, C.E.A, Turnbull, A, Ugochukwu, E, Papagrigoriou, E, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, Doyle, D.A. | | Deposit date: | 2006-07-03 | | Release date: | 2006-07-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Pick1 and Other Pdz Domains Obtained with the Help of Self-Binding C-Terminal Extensions.
Protein Sci., 16, 2007
|
|
2IWN
 
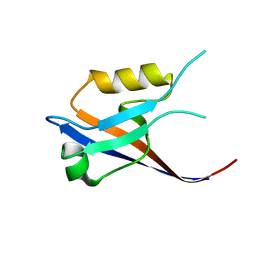 | | 3rd PDZ domain of Multiple PDZ Domain Protein MPDZ | | Descriptor: | MULTIPLE PDZ DOMAIN PROTEIN | | Authors: | Elkins, J.M, Gileadi, C, Savitsky, P, Berridge, G, Smee, C.E.A, Pike, A.C.W, Papagrigoriou, E, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, Doyle, D.A. | | Deposit date: | 2006-07-03 | | Release date: | 2006-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of Pick1 and Other Pdz Domains Obtained with the Help of Self-Binding C-Terminal Extension.
Protein Sci., 16, 2007
|
|
6JFP
 
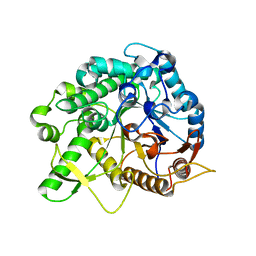 | | Crystal structure of the beta-glucosidase Bgl15 | | Descriptor: | beta-D-glucopyranose, beta-glucosidase 15 | | Authors: | Xie, W, Chen, R. | | Deposit date: | 2019-02-11 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Engineering of beta-Glucosidase Bgl15 with Simultaneously Enhanced Glucose Tolerance and Thermostability To Improve Its Performance in High-Solid Cellulose Hydrolysis.
J.Agric.Food Chem., 68, 2020
|
|
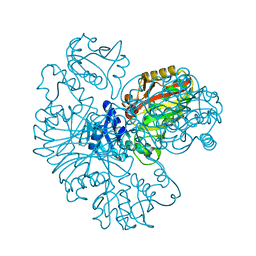
6IOJ
 
 | |
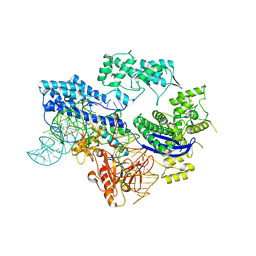
8HJ4
 
 | |
5Z1T
 
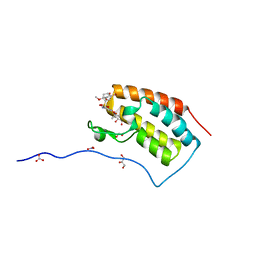 | | Crystal Structure Analysis of the BRD4(1) | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N-(6-hydroxy-2,2-dimethyl-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-7-yl)-2-methoxybenzene-1-sulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Xiang, Q, Song, M, Wang, C. | | Deposit date: | 2017-12-28 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Y08060: A Selective BET Inhibitor for Treatment of Prostate Cancer.
Acs Med.Chem.Lett., 9, 2018
|
|
5Z3W
 
 | | Malate dehydrogenase binds silver at C113 | | Descriptor: | Malate dehydrogenase, SILVER ION | | Authors: | Wang, H, Wang, M, Sun, H. | | Deposit date: | 2018-01-09 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Atomic differentiation of silver binding preference in protein targets: Escherichia coli malate dehydrogenase as a paradigm
Chem Sci, 2020
|
|
6J73
 
 | | Crystal structure of IniA from Mycobacterium smegmatis | | Descriptor: | Isoniazid inducible gene protein IniA | | Authors: | Wang, M.F, Guo, X.Y, Hu, J.J, Li, J, Rao, Z.H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.211 Å) | | Cite: | Mycobacterial dynamin-like protein IniA mediates membrane fission.
Nat Commun, 10, 2019
|
|
2LY7
 
 | | B-flap domain of RNA polymerase (B. subtilis) | | Descriptor: | DNA-directed RNA polymerase subunit beta | | Authors: | Mobli, M. | | Deposit date: | 2012-09-12 | | Release date: | 2014-03-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | RNA polymerase-induced remodelling of NusA produces a pause enhancement complex.
Nucleic Acids Res., 43, 2015
|
|
5Z1S
 
 | | Crystal Structure Analysis of the BRD4(1) | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-2-methoxy-N-(6-methoxy-2,2-dimethyl-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-7-yl)benzene-1-sulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Xiang, Q, Song, M, Wang, C. | | Deposit date: | 2017-12-28 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Y08060: A Selective BET Inhibitor for Treatment of Prostate Cancer.
Acs Med.Chem.Lett., 9, 2018
|
|
