7XTW
 
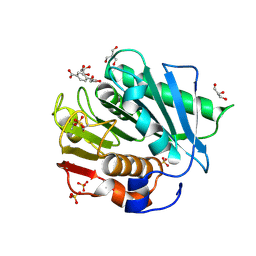 | | The structure of IsPETase in complex with MHET | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-hydroxyethyloxycarbonyl)benzoic acid, GLYCEROL, ... | | Authors: | Yang, Y, Jiang, P.C, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases.
Nat Commun, 14, 2023
|
|
7XTR
 
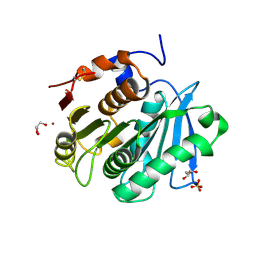 | | The apo structure of the engineered TfCut | | Descriptor: | GLYCEROL, LITHIUM ION, SULFATE ION, ... | | Authors: | Yang, Y, Jiang, P.C, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases.
Nat Commun, 14, 2023
|
|
7XTT
 
 | | The structure of engineered TfCut S130A in complex with MHET | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-hydroxyethyloxycarbonyl)benzoic acid, SODIUM ION, ... | | Authors: | Yang, Y, Jiang, P.C, Huang, J.W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases.
Nat Commun, 14, 2023
|
|
3V6G
 
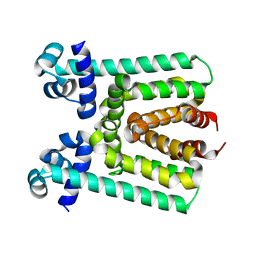 | | Crystal Structure of Transcriptional Regulator | | Descriptor: | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY DEOR-FAMILY) | | Authors: | Do, S.V, Bolla, J.R, Chen, X, Yu, E.W. | | Deposit date: | 2011-12-19 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | Structural and functional analysis of the transcriptional regulator Rv3066 of Mycobacterium tuberculosis.
Nucleic Acids Res., 40, 2012
|
|
7XS2
 
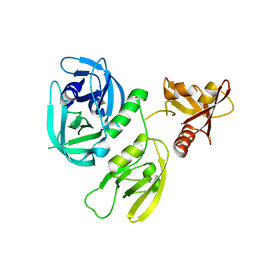 | | Monomer structure of HtrA from Helicobacter pylori | | Descriptor: | Periplasmic serine endoprotease DegP-like | | Authors: | Cui, L, Liu, W. | | Deposit date: | 2022-05-12 | | Release date: | 2023-05-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures and solution conformations of HtrA from Helicobacter pylori reveal pH-dependent oligomeric conversion and conformational rearrangements.
Int.J.Biol.Macromol., 243, 2023
|
|
7XS0
 
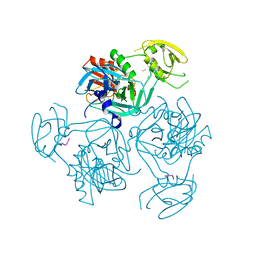 | | Trimer structure of HtrA from Helicobacter pylori bound with a tripeptide | | Descriptor: | Periplasmic serine endoprotease DegP-like, UNK-UNK-UNK | | Authors: | Cui, L, Liu, W. | | Deposit date: | 2022-05-12 | | Release date: | 2023-05-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structures and solution conformations of HtrA from Helicobacter pylori reveal pH-dependent oligomeric conversion and conformational rearrangements.
Int.J.Biol.Macromol., 243, 2023
|
|
3V78
 
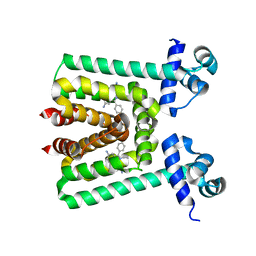 | | Crystal Structure of Transcriptional Regulator | | Descriptor: | ETHIDIUM, PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY DEOR-FAMILY) | | Authors: | Do, S.V, Bolla, J.R, Chen, X, Yu, E.W. | | Deposit date: | 2011-12-20 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural and functional analysis of the transcriptional regulator Rv3066 of Mycobacterium tuberculosis.
Nucleic Acids Res., 40, 2012
|
|
2LUQ
 
 | |
7EAX
 
 | | Crystal complex of p53-V272M and antimony ion | | Descriptor: | ANTIMONY (III) ION, Cellular tumor antigen p53, ZINC ION | | Authors: | Lu, M, Tang, Y. | | Deposit date: | 2021-03-08 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Repurposing antiparasitic antimonials to noncovalently rescue temperature-sensitive p53 mutations.
Cell Rep, 39, 2022
|
|
7YQG
 
 | |
7ZK5
 
 | | ABCB1 L335C mutant (mABCB1) in the outward facing state bound to AAC | | Descriptor: | (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent translocase ABCB1, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7ZK6
 
 | | ABCB1 L335C mutant (mABCB1) in the outward facing state bound to 2 molecules of AAC | | Descriptor: | (4S,11S,18S)-4-[[(2,4-dinitrophenyl)disulfanyl]methyl]-11,18-dimethyl-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7ZK4
 
 | | The ABCB1 L335C mutant (mABCB1) in the outward facing state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent translocase ABCB1, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7ZKB
 
 | | ABCB1 V978C mutant (mABCB1) in the inward facing state | | Descriptor: | (4S,11S,18S)-4-[[(2,4-dinitrophenyl)disulfanyl]methyl]-11,18-dimethyl-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ATP-dependent translocase ABCB1 | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7ZK8
 
 | | ABCB1 L971C mutant (mABCB1) in the outward facing state bound to AAC | | Descriptor: | (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent translocase ABCB1, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7CL0
 
 | | Crystal structure of human SIRT6 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NAD-dependent protein deacetylase sirtuin-6, ... | | Authors: | Song, K, Zhang, J. | | Deposit date: | 2020-07-20 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Reply to: Binding site for MDL-801 on SIRT6.
Nat.Chem.Biol., 17, 2021
|
|
7CL1
 
 | | Human SIRT6 in complex with allosteric activator MDL-801 (3.2A) | | Descriptor: | 5-[[3,5-bis(chloranyl)phenyl]sulfonylamino]-2-[(5-bromanyl-4-fluoranyl-2-methyl-phenyl)sulfamoyl]benzoic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Song, K, Zhang, J. | | Deposit date: | 2020-07-20 | | Release date: | 2021-02-24 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Reply to: Binding site for MDL-801 on SIRT6.
Nat.Chem.Biol., 17, 2021
|
|
7YQB
 
 | |
7WX2
 
 | | CBP-BrD complexed with NEO2734 | | Descriptor: | 1,3-dimethyl-5-[2-(oxan-4-yl)-3-[2-(trifluoromethyloxy)ethyl]benzimidazol-5-yl]pyridin-2-one, CREB-binding protein | | Authors: | Zeng, L, Lei, J.D. | | Deposit date: | 2022-02-14 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Targeting CDCP1 gene transcription coactivated by BRD4 and CBP/p300 in castration-resistant prostate cancer.
Oncogene, 41, 2022
|
|
7WWZ
 
 | | BRD4-BD1 complexed with NEO2734 | | Descriptor: | 1,3-dimethyl-5-[2-(oxan-4-yl)-3-[2-(trifluoromethyloxy)ethyl]benzimidazol-5-yl]pyridin-2-one, Isoform C of Bromodomain-containing protein 4 | | Authors: | Zeng, L, Lei, J.D. | | Deposit date: | 2022-02-14 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Targeting CDCP1 gene transcription coactivated by BRD4 and CBP/p300 in castration-resistant prostate cancer.
Oncogene, 41, 2022
|
|
4IGP
 
 | | Histone H3 Lysine 4 Demethylating Rice JMJ703 apo enzyme | | Descriptor: | FE (III) ION, Os05g0196500 protein | | Authors: | Chen, Q.F, Chen, X.S, Wang, Q, Zhang, F.B, Lou, Z.Y, Zhang, Q.F, Zhou, D.X. | | Deposit date: | 2012-12-17 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structural basis of a histone H3 lysine 4 demethylase required for stem elongation in rice.
PLoS Genet., 9, 2013
|
|
4IGQ
 
 | | Histone H3 Lysine 4 Demethylating Rice JMJ703 in complex with methylated H3K4 substrate | | Descriptor: | FE (III) ION, N-OXALYLGLYCINE, Os05g0196500 protein, ... | | Authors: | Chen, Q.F, Chen, X.S, Wang, Q, Zhang, F.B, Lou, Z.Y, Zhang, Q.F, Zhou, D.X. | | Deposit date: | 2012-12-17 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of a histone H3 lysine 4 demethylase required for stem elongation in rice.
PLoS Genet., 9, 2013
|
|
5XLM
 
 | | Monomer form of M.tuberculosis PknI sensor domain | | Descriptor: | Serine/threonine-protein kinase PknI | | Authors: | Rao, Z, Yan, Q. | | Deposit date: | 2017-05-10 | | Release date: | 2018-05-16 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insight into the Activation of PknI Kinase from M. tuberculosis via Dimerization of the Extracellular Sensor Domain.
Structure, 25, 2017
|
|
1WWN
 
 | | NMR Solution Structure of BmK-betaIT, an Excitatory Scorpion Toxin from Buthus martensi Karsch | | Descriptor: | Excitatory insect selective toxin 1 | | Authors: | Wu, H, Tong, X, Chen, X, Zhang, Q, Zheng, X, Zhang, N, Wu, G. | | Deposit date: | 2005-01-10 | | Release date: | 2006-01-17 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of BmK-betaIT, an excitatory scorpion beta-toxin without a 'hot spot' at the relevant position
Biochem.Biophys.Res.Commun., 349, 2006
|
|
2GA2
 
 | | h-MetAP2 complexed with A193400 | | Descriptor: | 5-BROMO-2-{[(4-CHLOROPHENYL)SULFONYL]AMINO}BENZOIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Park, C. | | Deposit date: | 2006-03-07 | | Release date: | 2007-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Development of sulfonamide compounds as potent methionine aminopeptidase type II inhibitors with antiproliferative properties.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
