2ICZ
 
 | | NMR Structures of the Expanded DNA 10bp xTGxTAxCxGCxAxGT:xACTxGCGxTAxCA | | Descriptor: | 5'-D(*(XAE)P*CP*TP*(XGA)P*CP*GP*(XTY)P*AP*(XCS)P*A)-3', 5'-D(*(XTY)P*GP*(XTY)P*AP*(XCS)P*(XGA)P*CP*(XAE)P*(XGA)P*T)-3' | | Authors: | Lynch, S.R. | | Deposit date: | 2006-09-13 | | Release date: | 2006-11-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Toward a Designed, Functioning Genetic System with Expanded-Size Base Pairs: Solution Structure of the Eight-Base xDNA Double Helix.
J.Am.Chem.Soc., 128, 2006
|
|
8HTV
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 3a | | Descriptor: | 1-(5,6-dihydrobenzo[b][1]benzazepin-11-yl)-2-sulfanyl-ethanone, 3C-like proteinase | | Authors: | Su, H.X, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2022-12-21 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Discovery and Mechanism Study of SARS-CoV-2 3C-like Protease Inhibitors with a New Reactive Group.
J.Med.Chem., 66, 2023
|
|
7FBD
 
 | | De novo design protein D53 with MBP tag | | Descriptor: | Maltodextrin-binding protein,De novo design protein D53 | | Authors: | Bin, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7FBB
 
 | | De novo design protein D12 with MBP tag | | Descriptor: | Maltodextrin-binding protein,de novo designed protein D12 | | Authors: | Bin, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7FBC
 
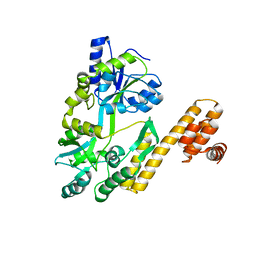 | | De novo design protein D22 with MBP tag | | Descriptor: | Maltodextrin-binding protein,De novo design protein D22 | | Authors: | Bin, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
8I71
 
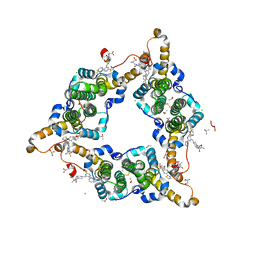 | | Hepatitis B virus core protein Y132A mutant in complex with Linvencorvir (RG7907), a Hepatitis B Virus (HBV) Core Protein Allosteric Modulator (CpAM) | | Descriptor: | 3-[(8~{a}~{S})-7-[[5-ethoxycarbonyl-4-(3-fluoranyl-2-methyl-phenyl)-2-(1,3-thiazol-2-yl)-1,4-dihydropyrimidin-6-yl]methyl]-3-oxidanylidene-5,6,8,8~{a}-tetrahydro-1~{H}-imidazo[1,5-a]pyrazin-2-yl]-2,2-dimethyl-propanoic acid, CHLORIDE ION, Capsid protein, ... | | Authors: | Zhou, Z, Xu, Z.H. | | Deposit date: | 2023-01-30 | | Release date: | 2023-03-22 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Linvencorvir (RG7907), a Hepatitis B Virus Core Protein Allosteric Modulator, for the Treatment of Chronic HBV Infection.
J.Med.Chem., 66, 2023
|
|
5FNV
 
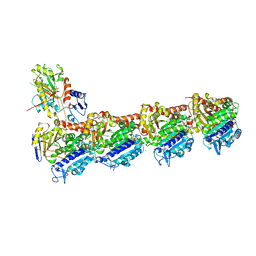 | | a new complex structure of tubulin with an alpha-beta unsaturated lactone | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, Y, Naismith, J, Zhu, X. | | Deposit date: | 2015-11-16 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Pironetin Reacts Covalently with Cysteine-316 of Alpha-Tubulin to Destabilize Microtubule.
Nat.Commun., 7, 2016
|
|
5HO4
 
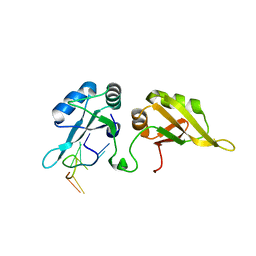 | | Crystal structure of hnRNPA2B1 in complex with 10-mer RNA | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1, RNA (5'-R(*AP*AP*GP*GP*AP*CP*UP*AP*GP*C)-3') | | Authors: | Wu, B.X, Su, S.C, Gan, J.H, Ma, J.B. | | Deposit date: | 2016-01-19 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1.
Nat Commun, 9, 2018
|
|
4K6J
 
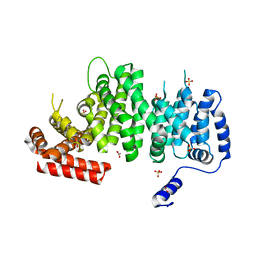 | | Human cohesin inhibitor WapL | | Descriptor: | ACETATE ION, SULFATE ION, Wings apart-like protein homolog | | Authors: | Tomchick, D.R, Yu, H, Ouyang, Z. | | Deposit date: | 2013-04-16 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6205 Å) | | Cite: | Structure of the human cohesin inhibitor Wapl.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6ENK
 
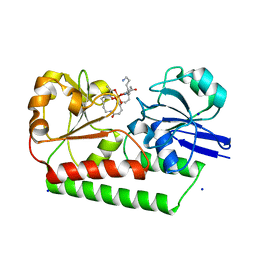 | | The X-ray crystal structure of DesE bound to desferrioxamine B | | Descriptor: | DesE, SODIUM ION, desferrioxamine B | | Authors: | Naismith, J.H, McMahon, S.A, Challis, G.L, Kadi, N, Oke, M, Liu, H, Carter, L.G, Johnson, K.A. | | Deposit date: | 2017-10-05 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Desferrioxamine biosynthesis: diverse hydroxamate assembly by substrate-tolerant acyl transferase DesC.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 373, 2018
|
|
2LWV
 
 | |
2LWT
 
 | |
2M0Q
 
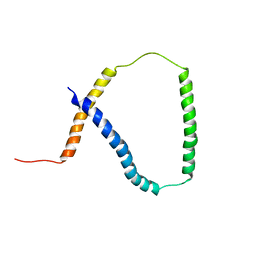 | | Solution NMR analysis of intact KCNE2 in detergent micelles demonstrate a straight transmembrane helix | | Descriptor: | Potassium voltage-gated channel subfamily E member 2 | | Authors: | Lai, C, Li, P, Chen, L, Zhang, L, Wu, F, Tian, C. | | Deposit date: | 2012-11-01 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Differential modulations of KCNQ1 by auxiliary proteins KCNE1 and KCNE2.
Sci Rep, 4, 2014
|
|
2LWU
 
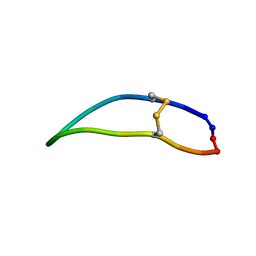 | |
2LWS
 
 | |
1XX2
 
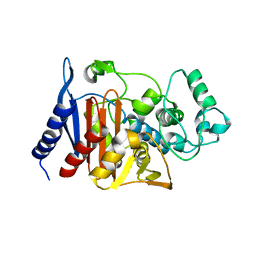 | | Refinement of P99 beta-lactamase from Enterobacter cloacae | | Descriptor: | Beta-lactamase | | Authors: | Knox, J.R, Sun, T. | | Deposit date: | 2004-11-03 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystallographic structure of a phosphonate derivative of the Enterobacter cloacae P99 cephalosporinase: mechanistic interpretation of a beta-lactamase transition-state analog.
Biochemistry, 33, 1994
|
|
2LWQ
 
 | |
7UIB
 
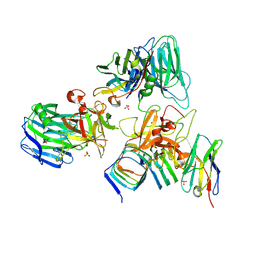 | | Crystal structure of BoNT/E receptor binding domain in complex with SV2, VHH, and sialic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-beta-neuraminic acid, ... | | Authors: | Liu, Z, Jin, R, Chen, P. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural basis for botulinum neurotoxin E recognition of synaptic vesicle protein 2.
Nat Commun, 14, 2023
|
|
7UIA
 
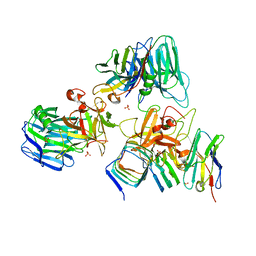 | | Crystal structure of BoNT/E receptor binding domain in complex with SV2 and VHH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Liu, Z, Jin, R, Chen, P. | | Deposit date: | 2022-03-28 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural basis for botulinum neurotoxin E recognition of synaptic vesicle protein 2.
Nat Commun, 14, 2023
|
|
2N2Y
 
 | |
2Q34
 
 | |
2Q35
 
 | |
2Q2X
 
 | |
6IMR
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | (1S)-1-[3-(1H-indol-3-yl)propyl]-6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-23 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6IM6
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 7-ethoxy-6-methoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-22 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
