4OLW
 
 | |
8YDC
 
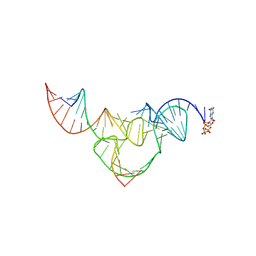 | | Crystal structure of a hammerhead ribozyme with pseudoknot | | Descriptor: | DNA/RNA (5'-R(*AP*CP*AP*UP*GP*UP*CP*U)-D(P*C)-R(P*UP*GP*GP*GP*A)-3'), GUANOSINE-5'-TRIPHOSPHATE, ribozyme strand | | Authors: | Liu, Y, Zhan, X. | | Deposit date: | 2024-02-20 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | The structure and catalytic mechanism of a pseudoknot-containing hammerhead ribozyme.
Nat Commun, 15, 2024
|
|
4OLU
 
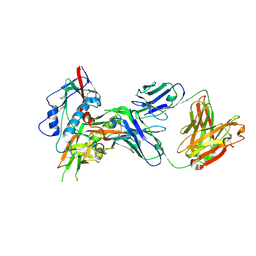 | | Crystal structure of antibody VRC07 in complex with clade A/E 93TH057 HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen binding fragment of heavy chain: Antibody VRC01, Antigen binding fragment of light chain: Antibody VRC01, ... | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2014-01-25 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Enhanced Potency of a Broadly Neutralizing HIV-1 Antibody In Vitro Improves Protection against Lentiviral Infection In Vivo.
J.Virol., 88, 2014
|
|
4OLX
 
 | |
8U3B
 
 | | Cryo-EM structure of E. coli NarL-transcription activation complex at 3.2A | | Descriptor: | DNA (69-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Kompaniiets, D, Wang, D. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis for transcription activation by the nitrate-responsive regulator NarL.
Nucleic Acids Res., 52, 2024
|
|
1UXD
 
 | | Fructose repressor DNA-binding domain, NMR, 34 structures | | Descriptor: | FRUCTOSE REPRESSOR | | Authors: | Penin, F, Geourjon, C, Montserret, R, Bockmann, A, Lesage, A, Yang, Y, Bonod-Bidaud, C, Cortay, J.C, Negre, D, Cozzone, A.J, Deleage, G. | | Deposit date: | 1996-12-26 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the DNA-binding domain of the fructose repressor from Escherichia coli by 1H and 15N NMR.
J.Mol.Biol., 270, 1997
|
|
4OLV
 
 | |
4OLY
 
 | |
1T2M
 
 | | Solution Structure Of The Pdz Domain Of AF-6 | | Descriptor: | AF-6 protein | | Authors: | Zhou, H, Wu, J.H, Xu, Y.Q, Huang, A.D, Shi, Y.Y. | | Deposit date: | 2004-04-22 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of AF-6 PDZ Domain and Its Interaction with the C-terminal Peptides from Neurexin and Bcr
J.Biol.Chem., 280, 2005
|
|
1UXC
 
 | | FRUCTOSE REPRESSOR DNA-BINDING DOMAIN, NMR, MINIMIZED STRUCTURE | | Descriptor: | FRUCTOSE REPRESSOR | | Authors: | Penin, F, Geourjon, C, Montserret, R, Bockmann, A, Lesage, A, Yang, Y, Bonod-Bidaud, C, Cortay, J.C, Negre, D, Cozzone, A.J, Deleage, G. | | Deposit date: | 1996-12-26 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the DNA-binding domain of the fructose repressor from Escherichia coli by 1H and 15N NMR.
J.Mol.Biol., 270, 1997
|
|
4I3R
 
 | | Crystal structure of the outer domain of HIV-1 gp120 in complex with VRC-PG04 space group P3221 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of VRC-PG04 Fab, Light chain of VRC-PG04 Fab, ... | | Authors: | Joyce, M.G, Biertumpfel, C, Nabel, G.J, Kwong, P.D. | | Deposit date: | 2012-11-26 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Outer Domain of HIV-1 gp120: Antigenic Optimization, Structural Malleability, and Crystal Structure with Antibody VRC-PG04.
J.Virol., 87, 2013
|
|
4I3S
 
 | | Crystal structure of the outer domain of HIV-1 gp120 in complex with VRC-PG04 space group P21 | | Descriptor: | CALCIUM ION, Heavy chain of VRC-PG04 Fab, Light chain of VRC-PG04 Fab, ... | | Authors: | Joyce, M.G, Biertumpfel, C, Nabel, G.J, Kwong, P.D. | | Deposit date: | 2012-11-26 | | Release date: | 2013-01-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Outer Domain of HIV-1 gp120: Antigenic Optimization, Structural Malleability, and Crystal Structure with Antibody VRC-PG04.
J.Virol., 87, 2013
|
|
8Z50
 
 | | Crystal structure of the ASF1-H3T-H4 complex | | Descriptor: | Histone H3.1t, Histone H4, Histone chaperone ASF1A | | Authors: | Xu, L. | | Deposit date: | 2024-04-18 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into instability of the nucleosome driven by histone variant H3T.
Biochem.Biophys.Res.Commun., 727, 2024
|
|
6O58
 
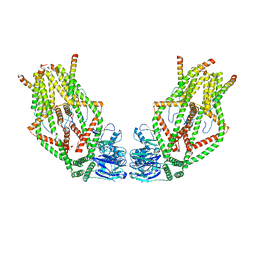 | | Human MCU-EMRE complex, dimer of channel | | Descriptor: | CALCIUM ION, Calcium uniporter protein, mitochondrial, ... | | Authors: | Wang, Y, Bai, X, Jiang, Y. | | Deposit date: | 2019-03-01 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Mechanism of EMRE-Dependent Gating of the Human Mitochondrial Calcium Uniporter.
Cell, 177, 2019
|
|
8WH4
 
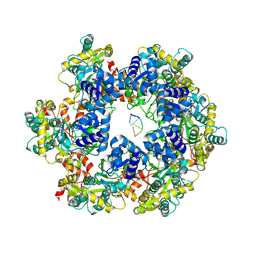 | | MPOX E5 hexamer ssDNA bound apo conformation | | Descriptor: | DNA (5'-D(P*CP*CP*CP*C)-3'), Uncoating factor OPG117 | | Authors: | Zhang, Z, Dong, C. | | Deposit date: | 2023-09-22 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Essential and multifunctional mpox virus E5 helicase-primase in double and single hexamer.
Sci Adv, 10, 2024
|
|
8WH3
 
 | | MPOX E5 hexamer apo form | | Descriptor: | Uncoating factor OPG117 | | Authors: | Zhang, Z, Dong, C. | | Deposit date: | 2023-09-22 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Essential and multifunctional mpox virus E5 helicase-primase in double and single hexamer.
Sci Adv, 10, 2024
|
|
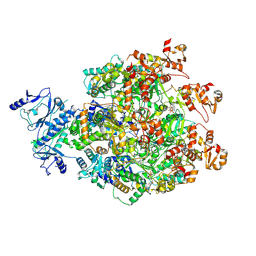
8WH6
 
 | |
8WGZ
 
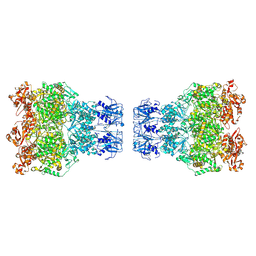 | | MPOX E5 double hexamer ssDNA bound conformation | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*C)-3'), Uncoating factor OPG117 | | Authors: | Zhang, Z, Dong, C. | | Deposit date: | 2023-09-22 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Essential and multifunctional mpox virus E5 helicase-primase in double and single hexamer.
Sci Adv, 10, 2024
|
|
8WGY
 
 | |
8WH0
 
 | | MPOX E5 hexamer ssDNA and AMP-PNP bound conformation | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*C)-3'), MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Zhang, Z, Dong, C. | | Deposit date: | 2023-09-22 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Essential and multifunctional mpox virus E5 helicase-primase in double and single hexamer.
Sci Adv, 10, 2024
|
|
1NYP
 
 | | 4th LIM domain of PINCH protein | | Descriptor: | PINCH protein, ZINC ION | | Authors: | Velyvis, A, Vaynberg, J, Vinogradova, O, Zhang, Y, Wu, C, Qin, J. | | Deposit date: | 2003-02-13 | | Release date: | 2003-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and functional insights into PINCH LIM4 domain-mediated integrin signaling
Nat.Struct.Biol., 10, 2003
|
|
4YRB
 
 | | mouse TDH mutant R180K with NAD+ bound | | Descriptor: | L-threonine 3-dehydrogenase, mitochondrial, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | He, C, Li, F. | | Deposit date: | 2015-03-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural insights on mouse l-threonine dehydrogenase: A regulatory role of Arg180 in catalysis
J.Struct.Biol., 192, 2015
|
|
4YR9
 
 | | mouse TDH with NAD+ bound | | Descriptor: | GLYCEROL, L-threonine 3-dehydrogenase, mitochondrial, ... | | Authors: | He, C, Li, F. | | Deposit date: | 2015-03-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights on mouse l-threonine dehydrogenase: A regulatory role of Arg180 in catalysis
J.Struct.Biol., 192, 2015
|
|
8H01
 
 | | SARS-CoV-2 Omicron BA.1 Spike glycoprotein in complex with rabbit monoclonal antibody 1H1 Fab in class 2 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-09-27 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanism of a rabbit monoclonal antibody broadly neutralizing SARS-CoV-2 variants.
Commun Biol, 6, 2023
|
|
8GZZ
 
 | | Local refinement of SARS-CoV-2 Omicron BA.1 Spike glycoprotein in complex with rabbit monoclonal antibody 1H1 Fab | | Descriptor: | Spike protein S1, rabbit monoclonal antibody 1H1 Fab heavy chain, rabbit monoclonal antibody 1H1 Fab light chain | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-09-27 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Mechanism of a rabbit monoclonal antibody broadly neutralizing SARS-CoV-2 variants.
Commun Biol, 6, 2023
|
|
