4R5Y
 
 | | The complex structure of Braf V600E kinase domain with a novel Braf inhibitor | | Descriptor: | 5-({(1R,1aS,6bR)-1-[5-(trifluoromethyl)-1H-benzimidazol-2-yl]-1a,6b-dihydro-1H-cyclopropa[b][1]benzofuran-5-yl}oxy)-3,4-dihydro-1,8-naphthyridin-2(1H)-one, Serine/threonine-protein kinase B-raf | | Authors: | Feng, Y, Peng, H, Zhang, Y, Liu, Y, Wei, M. | | Deposit date: | 2014-08-22 | | Release date: | 2016-02-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | BGB-283, a Novel RAF Kinase and EGFR Inhibitor, Displays Potent Antitumor Activity in BRAF-Mutated Colorectal Cancers.
Mol.Cancer Ther., 14, 2015
|
|
8U77
 
 | | Crystal structure of Taf14 in complex with Yng1 | | Descriptor: | Protein YNG1, Transcription initiation factor TFIID subunit 14 | | Authors: | Nguyen, M.C, Wei, P.C, Zhang, G.Y, Kutateladze, T.G. | | Deposit date: | 2023-09-14 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Molecular insight into interactions between the Taf14, Yng1 and Sas3 subunits of the NuA3 complex.
Nat Commun, 15, 2024
|
|
5Y1K
 
 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(2-chlorobenzyl)ureido)-N-hydroxy-4-methylpentanamide | | Descriptor: | (2S)-2-[(2-chlorophenyl)methylcarbamoylamino]-4-methyl-N-oxidanyl-pentanamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | Deposit date: | 2017-07-20 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Development of peptidomimetic hydroxamates as PfA-M1 and PfA-M17 dual inhibitors: Biological evaluation and structural characterization by cocrystallization
Chin.Chem.Lett., 33, 2022
|
|
5Y1Q
 
 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(3-chlorobenzyl)ureido)-N-hydroxy-4-methylpentanamide | | Descriptor: | (2S)-2-[(3-chlorophenyl)methylcarbamoylamino]-4-methyl-N-oxidanyl-pentanamide, M1 family aminopeptidase, MAGNESIUM ION, ... | | Authors: | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | Deposit date: | 2017-07-21 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Development of peptidomimetic hydroxamates as PfA-M1 and PfA-M17 dual inhibitors: Biological evaluation and structural characterization by cocrystallization
Chin.Chem.Lett., 33, 2022
|
|
5H57
 
 | | Ferredoxin III from maize root | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-3, chloroplastic | | Authors: | Kurisu, G, Hase, T. | | Deposit date: | 2016-11-04 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the isotype-specific interactions of ferredoxin and ferredoxin: NADP(+) oxidoreductase: an evolutionary switch between photosynthetic and heterotrophic assimilation
Photosyn. Res., 134, 2017
|
|
5H59
 
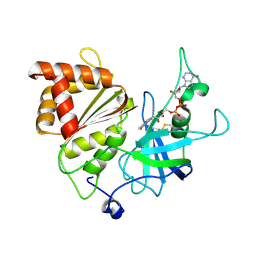 | | Ferredoxin-NADP+ reductase from maize root | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase | | Authors: | Kurisu, G, Hase, T. | | Deposit date: | 2016-11-04 | | Release date: | 2017-02-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for the isotype-specific interactions of ferredoxin and ferredoxin: NADP(+) oxidoreductase: an evolutionary switch between photosynthetic and heterotrophic assimilation
Photosyn. Res., 134, 2017
|
|
7YXV
 
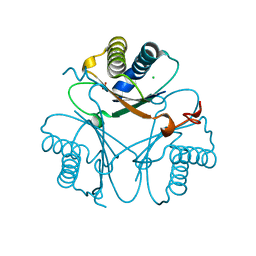 | |
4ER4
 
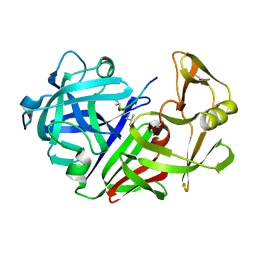 | | HIGH-RESOLUTION X-RAY ANALYSES OF RENIN INHIBITOR-ASPARTIC PROTEINASE COMPLEXES | | Descriptor: | ENDOTHIAPEPSIN, H-142 | | Authors: | Foundling, S.I, Watson, F.E, Szelke, M, Blundell, T.L. | | Deposit date: | 1991-01-05 | | Release date: | 1991-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High resolution X-ray analyses of renin inhibitor-aspartic proteinase complexes.
Nature, 327, 1987
|
|
5H5J
 
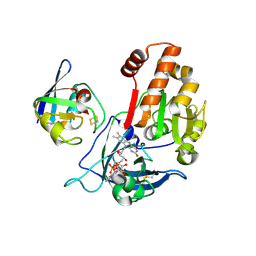 | | Complex between ferredoxin and ferredoxin-NADP+ reductase from maize root | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, ... | | Authors: | Kurisu, G, Hase, T. | | Deposit date: | 2016-11-05 | | Release date: | 2017-02-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the isotype-specific interactions of ferredoxin and ferredoxin: NADP(+) oxidoreductase: an evolutionary switch between photosynthetic and heterotrophic assimilation
Photosyn. Res., 134, 2017
|
|
4ER1
 
 | | THE ACTIVE SITE OF ASPARTIC PROTEINASES | | Descriptor: | ENDOTHIAPEPSIN, N-[(1R,2R,4R)-1-(cyclohexylmethyl)-2-hydroxy-6-methyl-4-{[(2R)-2-methylbutyl]carbamoyl}heptyl]-3-(1H-imidazol-3-ium-4-y l)-N~2~-[3-naphthalen-1-yl-2-(naphthalen-1-ylmethyl)propanoyl]-L-alaninamide | | Authors: | Quail, J.W, Cooper, J.B, Szelke, M, Blundell, T.L. | | Deposit date: | 1990-10-14 | | Release date: | 1991-01-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The active site of aspartic proteinases
FEBS Lett., 174, 1984
|
|
8V99
 
 | | GII.26 Leon 4509 norovirus protruding domain | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Capsid protein VP1, ... | | Authors: | Kher, G, Reese, T, Pancera, M, Hansman, G. | | Deposit date: | 2023-12-07 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Development of a broad-spectrum therapeutic Fc-nanobody for human noroviruses.
J.Virol., 98, 2024
|
|
8V98
 
 | | GII.24 Loreto 1972 norovirus protruding domain | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Capsid protein VP1 | | Authors: | Kher, G, Prewitt, A, Pancera, M, Hansman, G. | | Deposit date: | 2023-12-07 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Development of a broad-spectrum therapeutic Fc-nanobody for human noroviruses.
J.Virol., 98, 2024
|
|
8V9A
 
 | | GII.NA1 Loreto 1257 norovirus protruding domain | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein VP1 | | Authors: | Kher, G, Kim, I, Pancera, M, Hansman, G. | | Deposit date: | 2023-12-07 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Development of a broad-spectrum therapeutic Fc-nanobody for human noroviruses.
J.Virol., 98, 2024
|
|
8V97
 
 | | GII.17 CS-E1 norovirus protruding domain | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein VP1, DI(HYDROXYETHYL)ETHER | | Authors: | Kher, G, Prewitt, A, Pancera, M, Hansman, G. | | Deposit date: | 2023-12-07 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Development of a broad-spectrum therapeutic Fc-nanobody for human noroviruses.
J.Virol., 98, 2024
|
|
8V95
 
 | | GII.8 Amsterdam norovirus protruding domain | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein (Fragment) | | Authors: | Kher, G, Prewitt, A, Pancera, M, Hansman, G. | | Deposit date: | 2023-12-07 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Development of a broad-spectrum therapeutic Fc-nanobody for human noroviruses.
J.Virol., 98, 2024
|
|
8V96
 
 | | GII.14 M7 norovirus protruding domain | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein VP1 | | Authors: | Kher, G, Prewitt, A, Pancera, M, Hansman, G. | | Deposit date: | 2023-12-07 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Development of a broad-spectrum therapeutic Fc-nanobody for human noroviruses.
J.Virol., 98, 2024
|
|
5W05
 
 | | ANTI-TISSUE FACTOR ANTIBODY M59, A HUMANIZED VERSION OF 10H10 | | Descriptor: | DI(HYDROXYETHYL)ETHER, M59 FAB HEAVY CHAIN, M59 FAB LIGHT CHAIN | | Authors: | Teplyakov, A, Obmolova, G, Malia, T.J, Gilliland, G.L. | | Deposit date: | 2017-05-30 | | Release date: | 2017-06-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural insights into humanization of anti-tissue factor antibody 10H10.
MAbs, 10, 2018
|
|
3L5W
 
 | | Crystal structure of the complex between IL-13 and C836 FAB | | Descriptor: | C836 HEAVY CHAIN, C836 LIGHT CHAIN, GLYCEROL, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2009-12-22 | | Release date: | 2010-04-14 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human framework adaptation of a mouse anti-human IL-13 antibody.
J.Mol.Biol., 398, 2010
|
|
3L7F
 
 | | Structure of IL-13 antibody H2L6, A humanized variant OF C836 | | Descriptor: | CALCIUM ION, H2L6 HEAVY CHAIN, H2L6 LIGHT CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2009-12-28 | | Release date: | 2010-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human framework adaptation of a mouse anti-human IL-13 antibody.
J.Mol.Biol., 398, 2010
|
|
7TE7
 
 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with RAD1901 | | Descriptor: | (6R)-6-{2-[ethyl({4-[2-(ethylamino)ethyl]phenyl}methyl)amino]-4-methoxyphenyl}-5,6,7,8-tetrahydronaphthalen-2-ol, Estrogen receptor | | Authors: | Joiner, C, Hancock, G, Young, K, Hosfield, D.J, Greene, G.L, Fanning, S.W. | | Deposit date: | 2022-01-04 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Unconventional isoquinoline-based SERMs elicit fulvestrant-like transcriptional programs in ER+ breast cancer cells.
NPJ Breast Cancer, 8, 2022
|
|
1PYP
 
 | |
8WBG
 
 | |
8WBF
 
 | |
8WBH
 
 | |
4QSZ
 
 | |
