4R0M
 
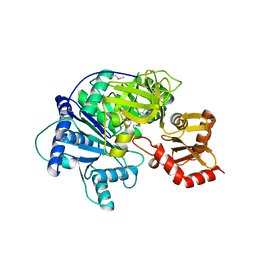 | | Structure of McyG A-PCP complexed with phenylalanyl-adenylate | | Descriptor: | ADENOSINE-5'-[PHENYLALANINYL-PHOSPHATE], McyG protein | | Authors: | Tan, X.F, Dai, Y.N, Zhou, K, Jiang, Y.L, Ren, Y.M, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2014-08-01 | | Release date: | 2015-04-15 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the adenylation-peptidyl carrier protein didomain of the Microcystis aeruginosa microcystin synthetase McyG.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4QSG
 
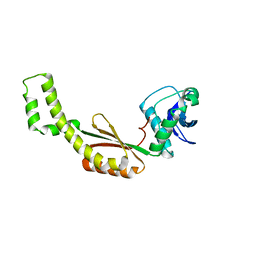 | | Crystal structure of gas vesicle protein GvpF from Microcystis aeruginosa | | Descriptor: | Gas vesicle protein | | Authors: | Xu, B.Y, Dai, Y.N, Zhou, K, Ren, Y.M, Liu, Y.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2014-07-03 | | Release date: | 2014-11-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the gas vesicle protein GvpF from the cyanobacterium Microcystis aeruginosa.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6J3Q
 
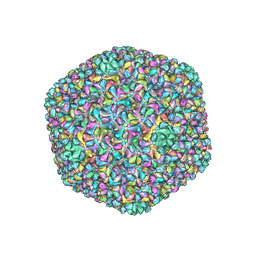 | | Capsid structure of a freshwater cyanophage Siphoviridae Mic1 | | Descriptor: | cement protein, major capsid protein | | Authors: | Jin, H, Jiang, Y.L, Yang, F, Zhang, J.T, Li, W.F, Zhou, K, Ju, J, Chen, Y, Zhou, C.Z. | | Deposit date: | 2019-01-05 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Capsid Structure of a Freshwater Cyanophage Siphoviridae Mic1.
Structure, 27, 2019
|
|
5WXZ
 
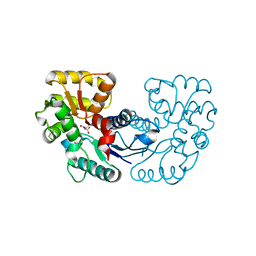 | | Crystal structure of Microcystis aeruginosa PCC 7806 aspartate racemase in complex with D-aspartate | | Descriptor: | D-ASPARTIC ACID, McyF | | Authors: | Cao, D.D, Zhou, K, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2017-01-09 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the catalysis and substrate specificity of cyanobacterial aspartate racemase McyF.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
5G06
 
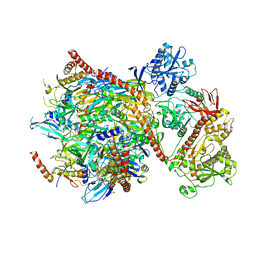 | | Cryo-EM structure of yeast cytoplasmic exosome | | Descriptor: | EXOSOME COMPLEX COMPONENT CSL4, EXOSOME COMPLEX COMPONENT MTR3, EXOSOME COMPLEX COMPONENT RRP4, ... | | Authors: | Liu, J.J, Niu, C.Y, Wu, Y, Tan, D, Wang, Y, Ye, M.D, Liu, Y, Zhao, W.W, Zhou, K, Liu, Q.S, Dai, J.B, Yang, X.R, Dong, M.Q, Huang, N, Wang, H.W. | | Deposit date: | 2016-03-17 | | Release date: | 2016-06-15 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryoem Structure of Yeast Cytoplasmic Exosome Complex.
Cell Res., 26, 2016
|
|
5IFG
 
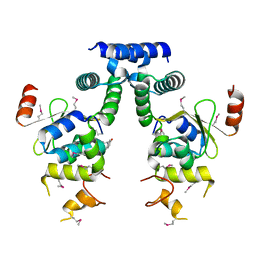 | | Crystal structure of HigA-HigB complex from E. Coli | | Descriptor: | Antitoxin HigA, mRNA interferase HigB | | Authors: | Yang, J.S, Zhou, K, Gao, z.Q, Liu, Q.S, Dong, Y.H. | | Deposit date: | 2016-02-26 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Structural insight into the E. coli HigBA complex
Biochem. Biophys. Res. Commun., 478, 2016
|
|
6J4A
 
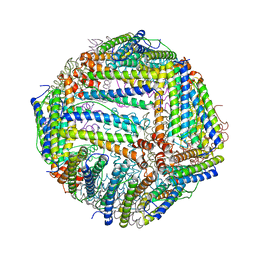 | |
6J4M
 
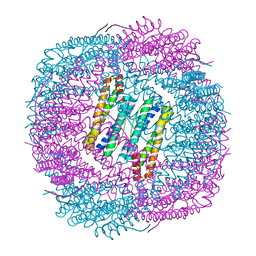 | | Thermal treated soybean seed H-2 ferritin | | Descriptor: | Ferritin, MAGNESIUM ION | | Authors: | Zhang, X, Zang, J, Chen, H, Zhou, K, Zhao, G. | | Deposit date: | 2019-01-09 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Thermostability of protein nanocages: the effect of natural extra peptide on the exterior surface.
Rsc Adv, 9, 2019
|
|
5YOX
 
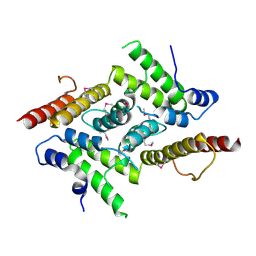 | | HD domain-containing protein YGK1(YGL101W) | | Descriptor: | HD domain-containing protein YGL101W, ZINC ION | | Authors: | Yang, J, Wang, F, Gao, Z, Zhou, K, Liu, Q. | | Deposit date: | 2017-10-31 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | HD domain-containing protein YGK1(YGL101W)
To Be Published
|
|
6JOB
 
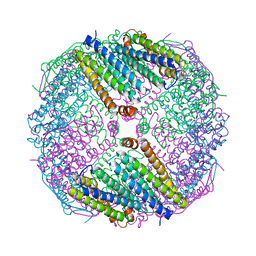 | | Ferritin variant with "GMG" motif | | Descriptor: | Ferritin heavy chain | | Authors: | Zheng, B.W, Zhou, K, Zhang, T, Lv, C, Wang, H, Zhao, G. | | Deposit date: | 2019-03-20 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | SOLUTION SCATTERING (2.93 Å), X-RAY DIFFRACTION | | Cite: | Self-assembly of protein nanocage into designed 2D and 3D networks by grafting amyloidogenic motifs on the exterior surfaces.
To Be Published
|
|
6TY9
 
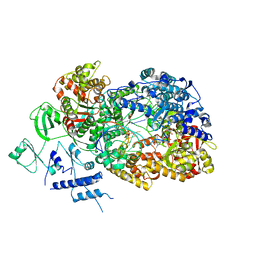 | | In situ structure of BmCPV RNA dependent RNA polymerase at initiation state | | Descriptor: | MAGNESIUM ION, Non-template RNA (5'-D(*(GTA))-R(P*GP*UP*AP*AP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3'), RNA-dependent RNA Polymerase, ... | | Authors: | Cui, Y.X, Zhang, Y.N, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2019-08-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Conservative transcription in three steps visualized in a double-stranded RNA virus.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6TZ1
 
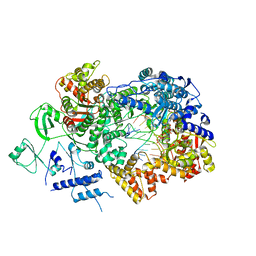 | | In situ structure of BmCPV RNA-dependent RNA polymerase at early-elongation state | | Descriptor: | Non-template RNA (5'-D(*(GTA))-R(P*GP*UP*A)-3'), RNA-dependent RNA Polymerase, Template RNA (5'-R(P*AP*GP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), ... | | Authors: | Cui, Y.X, Zhang, Y.N, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2019-08-09 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Conservative transcription in three steps visualized in a double-stranded RNA virus.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6TY8
 
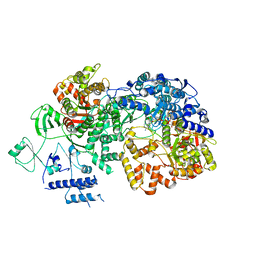 | | In situ structure of BmCPV RNA dependent RNA polymerase at quiescent state | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, RNA-dependent RNA Polymerase, Viral structural protein 4 | | Authors: | Cui, Y.X, Zhang, Y.N, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2019-08-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conservative transcription in three steps visualized in a double-stranded RNA virus.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6TZ2
 
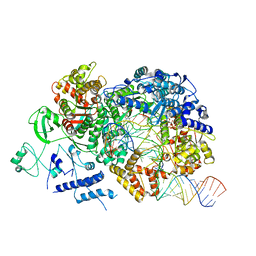 | | In situ structure of BmCPV RNA-dependent RNA polymerase at elongation state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Non-template RNA (36-MER), ... | | Authors: | Cui, Y.X, Zhang, Y.N, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2019-08-09 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conservative transcription in three steps visualized in a double-stranded RNA virus.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6TZ0
 
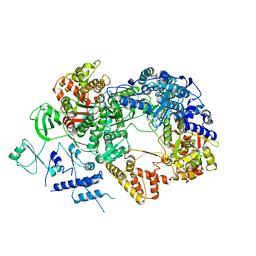 | | In situ structure of BmCPV RNA-dependent RNA polymerase at abortive state | | Descriptor: | RNA-dependent RNA Polymerase, Viral structural protein 4 | | Authors: | Cui, Y.X, Zhang, Y.N, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2019-08-09 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Conservative transcription in three steps visualized in a double-stranded RNA virus.
Nat.Struct.Mol.Biol., 26, 2019
|
|
3O05
 
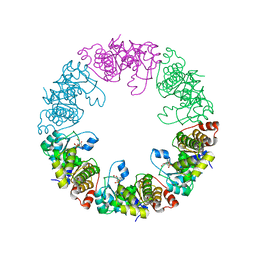 | | Crystal Structure of Yeast Pyridoxal 5-Phosphate Synthase Snz1 Complxed with Substrate PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Pyridoxine biosynthesis protein SNZ1 | | Authors: | Teng, Y.B, Zhang, X, He, Y.X, Hu, H.X, Zhou, C.Z. | | Deposit date: | 2010-07-19 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the catalytic mechanism of the yeast pyridoxal 5-phosphate synthase Snz1
Biochem.J., 432, 2010
|
|
3O06
 
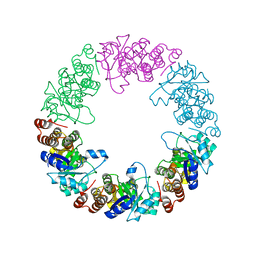 | |
3O07
 
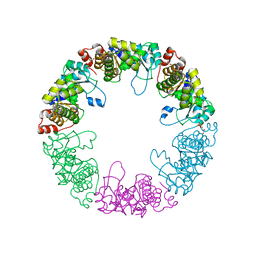 | | Crystal structure of yeast pyridoxal 5-phosphate synthase Snz1 complexed with substrate G3P | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, Pyridoxine biosynthesis protein SNZ1 | | Authors: | Teng, Y.B, Zhang, X, Hu, H.X, Zhou, C.Z. | | Deposit date: | 2010-07-19 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the catalytic mechanism of the yeast pyridoxal 5-phosphate synthase Snz1
Biochem.J., 432, 2010
|
|
6EQT
 
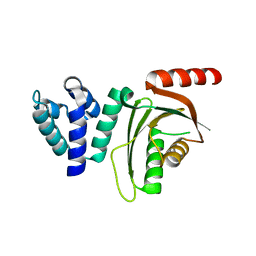 | |
4LMA
 
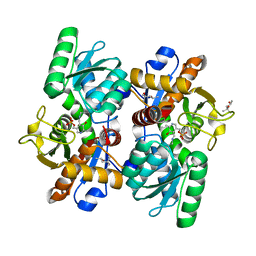 | |
4LMB
 
 | |
4N0S
 
 | | Complex of ERK2 with caffeic acid | | Descriptor: | CAFFEIC ACID, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Kurinov, I, Malakhova, M. | | Deposit date: | 2013-10-02 | | Release date: | 2014-08-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7992 Å) | | Cite: | Caffeic Acid Directly Targets ERK1/2 to Attenuate Solar UV-Induced Skin Carcinogenesis.
Cancer Prev Res (Phila), 7, 2014
|
|
6ECN
 
 | | HIV-1 CA 1/2-hexamer-EE | | Descriptor: | HIV-1 CA | | Authors: | Summers, B.J, Xiong, Y. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Modular HIV-1 Capsid Assemblies Reveal Diverse Host-Capsid Recognition Mechanisms.
Cell Host Microbe, 26, 2019
|
|
6EC2
 
 | | Structure of HIV-1 CA 1/3-hexamer | | Descriptor: | ACETATE ION, Capsid protein p24 | | Authors: | Summers, B.J, Xiong, Y. | | Deposit date: | 2018-08-07 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Modular HIV-1 Capsid Assemblies Reveal Diverse Host-Capsid Recognition Mechanisms.
Cell Host Microbe, 26, 2019
|
|
6ECO
 
 | | Hexamer-2-Foldon HIV-1 capsid platform | | Descriptor: | HIV-1 capsid platform protein | | Authors: | Summers, B.J, Xiong, Y. | | Deposit date: | 2018-08-08 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Modular HIV-1 Capsid Assemblies Reveal Diverse Host-Capsid Recognition Mechanisms.
Cell Host Microbe, 26, 2019
|
|
