1G9C
 
 | |
1G9B
 
 | |
1G9A
 
 | |
5D7V
 
 | | Crystal structure of PTK6 kinase domain | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein-tyrosine kinase 6 | | Authors: | Thakur, M.K, Birudukota, S, Swaminathan, S, Tyagi, R, Gosu, R. | | Deposit date: | 2015-08-14 | | Release date: | 2016-08-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of the kinase domain of human protein tyrosine kinase 6 (PTK6) at 2.33 angstrom resolution
Biochem.Biophys.Res.Commun., 478, 2016
|
|
4MYM
 
 | | Crystal structure of a glyoxalase/ bleomycin resistance protein/ dioxygenase from Nocardioides | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glyoxalase/bleomycin resistance protein/dioxygenase | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, AL Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-09-27 | | Release date: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a glyoxalase/ bleomycin resistance protein/ dioxygenase from Nocardioides.
To be Published
|
|
1CCD
 
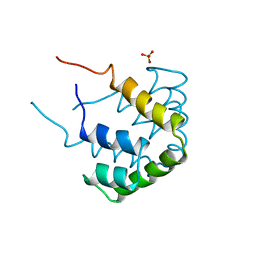 | | REFINED STRUCTURE OF RAT CLARA CELL 17 KDA PROTEIN AT 3.0 ANGSTROMS RESOLUTION | | Descriptor: | CLARA CELL 17 kD PROTEIN, SULFATE ION | | Authors: | Umland, T.C, Swaminathan, S, Furey, W, Singh, G, Pletcher, J, Sax, M. | | Deposit date: | 1991-09-17 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Refined structure of rat Clara cell 17 kDa protein at 3.0 A resolution.
J.Mol.Biol., 224, 1992
|
|
4QB5
 
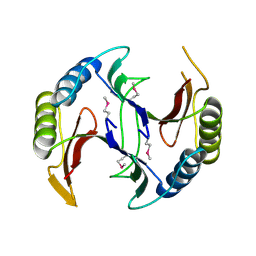 | | Crystal structure of a glyoxalase/bleomycin resistance protein from Albidiferax ferrireducens T118 | | Descriptor: | 1,2-ETHANEDIOL, Glyoxalase/bleomycin resistance protein/dioxygenase, SULFATE ION | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-05-06 | | Release date: | 2014-07-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a glyoxalase/bleomycin resistance protein from Albidiferax ferrireducens T118
To be Published
|
|
4M1A
 
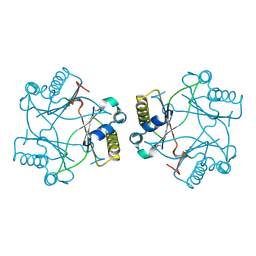 | | Crystal structure of a Domain of unknown function (DUF1904) from Sebaldella termitidis ATCC 33386 | | Descriptor: | Hypothetical protein | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-02 | | Release date: | 2013-08-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a Domain of unknown function (DUF1904) from Sebaldellatermitidis ATCC 33386
To be Published
|
|
4LK5
 
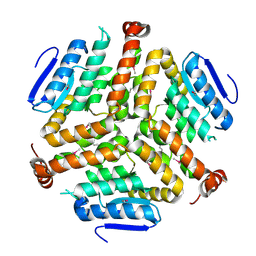 | | Crystal structure of a enoyl-CoA hydratase from Mycobacterium avium subsp. paratuberculosis K-10 | | Descriptor: | enoyl-CoA hydratase | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-06 | | Release date: | 2013-07-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a enoyl-CoA hydratase from Mycobacterium avium subsp. paratuberculosis K-10
To be Published
|
|
4LKB
 
 | | Crystal structure of a putative 4-Oxalocrotonate Tautomerase from Nostoc sp. PCC 7120 | | Descriptor: | GLYCEROL, SULFATE ION, hypothetical protein alr4568/putative 4-Oxalocrotonate Tautomerase | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-07 | | Release date: | 2013-07-24 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of a putative 4-Oxalocrotonate Tautomerase from Nostoc sp. PCC 7120
To be Published
|
|
1AF9
 
 | | TETANUS NEUROTOXIN C FRAGMENT | | Descriptor: | TETANUS NEUROTOXIN | | Authors: | Umland, T.C, Wingert, L, Swaminathan, S, Furey, W.F, Schmidt, J.J, Sax, M. | | Deposit date: | 1997-03-24 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the receptor binding fragment HC of tetanus neurotoxin.
Nat.Struct.Biol., 4, 1997
|
|
5YJI
 
 | | Co-crystal structure of Mouse Nicotinamide N-methyltransferase (NNMT) with small molecule analog of Nicotinamide | | Descriptor: | 6-methoxy-1-methyl-2H-pyridine-3-carboxamide, Nicotinamide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Birudukota, S, Swaminathan, S, Thakur, M.K, Parveen, R, Kandan, S, Hallur, M.S, Rajagopal, S, Ruf, S, Dhakshinamoorthy, S, Kannt, A, Gosu, R. | | Deposit date: | 2017-10-10 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A small molecule inhibitor of Nicotinamide N-methyltransferase for the treatment of metabolic disorders.
Sci Rep, 8, 2018
|
|
5BUX
 
 | |
5BUW
 
 | |
5BUY
 
 | |
1CT5
 
 | | CRYSTAL STRUCTURE OF YEAST HYPOTHETICAL PROTEIN YBL036C-SELENOMET CRYSTAL | | Descriptor: | PROTEIN (YEAST HYPOTHETICAL PROTEIN, SELENOMET), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Eswaramoorthy, S, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 1999-08-18 | | Release date: | 1999-09-02 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a yeast hypothetical protein selected by a structural genomics approach.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4ZKT
 
 | |
1F1M
 
 | | CRYSTAL STRUCTURE OF OUTER SURFACE PROTEIN C (OSPC) | | Descriptor: | OUTER SURFACE PROTEIN C, ZINC ION | | Authors: | Kumaran, D, Eswaramoorthy, S, Dunn, J.J, Swaminathan, S. | | Deposit date: | 2000-05-19 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of outer surface protein C (OspC) from the Lyme disease spirochete, Borrelia burgdorferi.
EMBO J., 20, 2001
|
|
1NJR
 
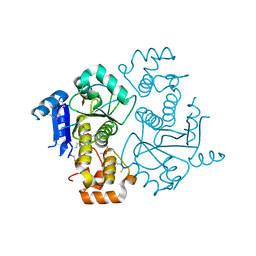 | | Crystal structure of yeast ymx7, an ADP-ribose-1''-monophosphatase | | Descriptor: | 32.1 kDa protein in ADH3-RCA1 intergenic region, Xylitol | | Authors: | Kumaran, D, Eswaramoorthy, S, Studier, F.W, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-01-02 | | Release date: | 2004-08-17 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of ADP-ribose-1''-monophosphatase (Appr-1''-pase), a ubiquitous cellular processing enzyme
Protein Sci., 14, 2005
|
|
1NKQ
 
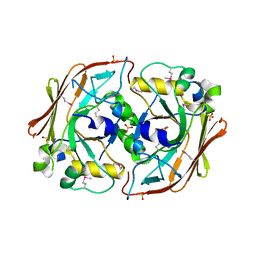 | | Crystal structure of yeast ynq8, a fumarylacetoacetate hydrolase family protein | | Descriptor: | ACETIC ACID, CALCIUM ION, Hypothetical 28.8 kDa protein in PSD1-SKO1 intergenic region, ... | | Authors: | Eswaramoorthy, S, Kumaran, D, Daniels, B, Studier, F.W, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-01-03 | | Release date: | 2004-06-15 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crtystal Structure of Yeast Hypothetical Protein YNQ8_YEAST
To be Published
|
|
1QDF
 
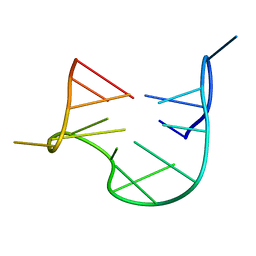 | | THE NMR STUDY OF DNA QUADRUPLEX STRUCTURE, APTAMER (15MER) DNA | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3') | | Authors: | Marathias, V.M, Wang, K.Y, Kumar, S, Swaminathan, S, Bolton, P.H. | | Deposit date: | 1996-04-11 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Determination of the number and location of the manganese binding sites of DNA quadruplexes in solution by EPR and NMR in the presence and absence of thrombin.
J.Mol.Biol., 260, 1996
|
|
1QDI
 
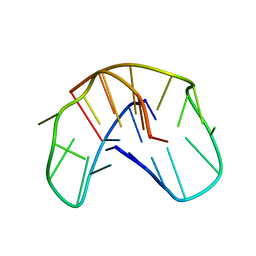 | | THE NMR STUDY OF DNA QUADRUPLEX STRUCTURE, (12MER) DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3') | | Authors: | Marathias, V.M, Wang, K.Y, Kumar, S, Swaminathan, S, Bolton, P.H. | | Deposit date: | 1996-04-11 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Determination of the number and location of the manganese binding sites of DNA quadruplexes in solution by EPR and NMR in the presence and absence of thrombin.
J.Mol.Biol., 260, 1996
|
|
1QDH
 
 | | THE NMR STUDY OF DNA QUADRUPLEX STRUCTURE, APTAMER (15MER) DNA | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3'), MANGANESE (II) ION | | Authors: | Marathias, V.M, Wang, K.Y, Kumar, S, Swaminathan, S, Bolton, P.H. | | Deposit date: | 1996-04-11 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Determination of the number and location of the manganese binding sites of DNA quadruplexes in solution by EPR and NMR in the presence and absence of thrombin.
J.Mol.Biol., 260, 1996
|
|
1QDK
 
 | | THE NMR STUDY OF DNA QUADRUPLEX STRUCTURE, (12MER) DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3'), MANGANESE (II) ION | | Authors: | Marathias, V.M, Wang, K.Y, Kumar, S, Swaminathan, S, Bolton, P.H. | | Deposit date: | 1996-04-11 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Determination of the number and location of the manganese binding sites of DNA quadruplexes in solution by EPR and NMR in the presence and absence of thrombin.
J.Mol.Biol., 260, 1996
|
|
1F89
 
 | | Crystal structure of Saccharomyces cerevisiae Nit3, a member of branch 10 of the nitrilase superfamily | | Descriptor: | 32.5 KDA PROTEIN YLR351C | | Authors: | Kumaran, D, Eswaramoorthy, S, Studier, F.W, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2000-06-29 | | Release date: | 2001-10-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a putative CN hydrolase from yeast
Proteins, 52, 2003
|
|
