1WSG
 
 | | Co-crystal structure of E.coli RNase HI active site mutant (E48A/D134N*) with Mn2+ | | Descriptor: | MANGANESE (II) ION, Ribonuclease HI | | Authors: | Tsunaka, Y, Takano, K, Matsumura, H, Yamagata, Y, Kanaya, S. | | Deposit date: | 2004-11-05 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of Single Mn(2+) Binding Sites Required for Activation of the Mutant Proteins of E.coli RNase HI at Glu48 and/or Asp134 by X-ray Crystallography
J.Mol.Biol., 345, 2005
|
|
5B4W
 
 | | Crystal structure of Plexin inhibitor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-B1, Synthesized cyclic peptide | | Authors: | Matsunaga, Y, Kitago, Y, Arimori, T, Takagi, J. | | Deposit date: | 2016-04-19 | | Release date: | 2016-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Allosteric Inhibition of a Semaphorin 4D Receptor Plexin B1 by a High-Affinity Macrocyclic Peptide
Cell Chem Biol, 23, 2016
|
|
2CV5
 
 | | Crystal structure of human nucleosome core particle | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A.a, ... | | Authors: | Tsunaka, Y, Kajimura, N, Tate, S, Morikawa, K. | | Deposit date: | 2005-05-31 | | Release date: | 2005-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Alteration of the nucleosomal DNA path in the crystal structure of a human nucleosome core particle
Nucleic Acids Res., 33, 2005
|
|
8BCM
 
 | |
9C96
 
 | | Cryo-EM structure of TAP binding protein related (TAPBPR) in complex with HLA-A*02:01 bound to a suboptimal peptide. | | Descriptor: | Beta-2-microglobulin, LYS-ILE-LEU-GLY-PHE-VAL, MHC class I antigen, ... | | Authors: | Pumroy, R.P, Mallik, L, Sun, Y, Moiseenkova-Bell, Y.V, Sgourakis, N.G. | | Deposit date: | 2024-06-13 | | Release date: | 2025-01-22 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | CryoEM structure of an MHC-I/TAPBPR peptide-bound intermediate reveals the mechanism of antigen proofreading.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
6URG
 
 | | Cryo-EM structure of human CPSF160-WDR33-CPSF30-CPSF100 PIM complex | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 2, Cleavage and polyadenylation specificity factor subunit 4, ... | | Authors: | Sun, Y, Zhang, Y, Walz, T, Tong, L. | | Deposit date: | 2019-10-23 | | Release date: | 2019-11-27 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural Insights into the Human Pre-mRNA 3'-End Processing Machinery.
Mol.Cell, 77, 2020
|
|
6URO
 
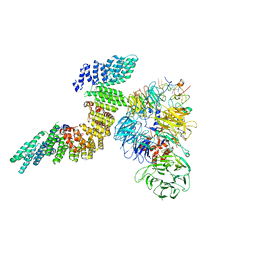 | | Cryo-EM structure of human CPSF160-WDR33-CPSF30-PAS RNA-CstF77 complex | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 4, Cleavage stimulation factor subunit 3, ... | | Authors: | Sun, Y, Zhang, Y, Walz, T, Tong, L. | | Deposit date: | 2019-10-23 | | Release date: | 2019-11-27 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Insights into the Human Pre-mRNA 3'-End Processing Machinery.
Mol.Cell, 77, 2020
|
|
4M7E
 
 | | Structural insight into BL-induced activation of the BRI1-BAK1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1, ... | | Authors: | Chai, J, Han, Z, Sun, Y. | | Deposit date: | 2013-08-12 | | Release date: | 2013-10-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.602 Å) | | Cite: | Structural insight into BL-induced activation of the BRI1-BAK1 complex
To be Published
|
|
4MN8
 
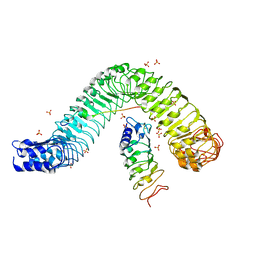 | | Crystal structure of flg22 in complex with the FLS2 and BAK1 ectodomains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1, ... | | Authors: | Chai, J, Sun, Y, Han, Z. | | Deposit date: | 2013-09-10 | | Release date: | 2013-12-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.062 Å) | | Cite: | Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex.
Science, 342, 2013
|
|
6NN3
 
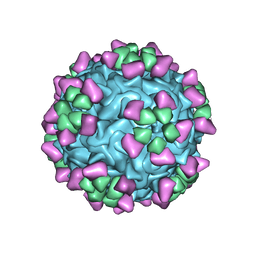 | | Structure of parvovirus B19 decorated with Fab molecules from a human antibody | | Descriptor: | Fab monoclonal antibody 860-55D, heavy chain, light chain, ... | | Authors: | Rossmann, M.G, Sun, Y, Klose, T, Liu, Y. | | Deposit date: | 2019-01-14 | | Release date: | 2019-02-13 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structure of Parvovirus B19 Decorated by Fabs from a Human Antibody.
J. Virol., 93, 2019
|
|
8ISS
 
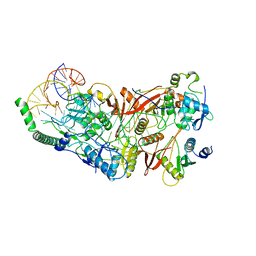 | | Cryo-EM structure of wild-type human tRNA Splicing Endonuclease Complex bound to pre-tRNA-ARG at 3.19 A resolution | | Descriptor: | MAGNESIUM ION, RNA (88-MER), tRNA-splicing endonuclease subunit Sen15, ... | | Authors: | Sun, Y, Zhang, Y, Yuan, L, Han, Y. | | Deposit date: | 2023-03-21 | | Release date: | 2023-10-18 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Recognition and cleavage mechanism of intron-containing pre-tRNA by human TSEN endonuclease complex.
Nat Commun, 14, 2023
|
|
8WZ4
 
 | | Cryo-EM structure of prefusion-stabilized RSV F (DS-Cav1 strain: A2) in complex with nAb 5B11 (localized refinement) | | Descriptor: | 5B11 Fab Heavy Chain, 5B11 Fab Light Chain, RSV Fusion glycoprotein | | Authors: | Liu, L, Sun, H, Sun, Y, Zheng, Q, Li, S, Zheng, Z, Xia, N. | | Deposit date: | 2023-11-01 | | Release date: | 2024-11-06 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | A potent broad-spectrum neutralizing antibody targeting a conserved region of the prefusion RSV F protein.
Nat Commun, 15, 2024
|
|
8WZ3
 
 | | Cryo-EM structure of prefusion-stabilized RSV F (DS-Cav1 strain: A2) in complex with nAb 5B11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5B11 Fab Heavy Chain, 5B11 Fab Light Chain, ... | | Authors: | Liu, L, Sun, H, Sun, Y, Zheng, Q, Li, S, Zheng, Z, Xia, N. | | Deposit date: | 2023-11-01 | | Release date: | 2024-11-13 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | A potent broad-spectrum neutralizing antibody targeting a conserved region of the prefusion RSV F protein.
Nat Commun, 15, 2024
|
|
8WZ5
 
 | | Cryo-EM structure of prefusion-stabilized RSV F (DS-Cav1 sc9-10 strain: B18537) in complex with humanized nAb 5B11 | | Descriptor: | 5B11 Fab Heavy Chain, 5B11 Fab Light Chain, RSV Fusion glycoprotein | | Authors: | Liu, L, Sun, H, Sun, Y, Zheng, Q, Li, S, Zheng, Z, Xia, N. | | Deposit date: | 2023-11-01 | | Release date: | 2024-11-13 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | A potent broad-spectrum neutralizing antibody targeting a conserved region of the prefusion RSV F protein.
Nat Commun, 15, 2024
|
|
8WZE
 
 | | Cryo-EM structure of prefusion-stabilized RSV F (DS-Cav1 sc9-10 strain: B18537) in complex with humanized nAb 5B11 (localized refinement) | | Descriptor: | 5B11 Fab Heavy Chain, 5B11 Fab Light Chain, RSV pre-fusion glycoprotein | | Authors: | Liu, L, Sun, H, Sun, Y, Zheng, Q, Li, S, Zheng, Z, Xia, N. | | Deposit date: | 2023-11-02 | | Release date: | 2024-11-13 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | A potent broad-spectrum neutralizing antibody targeting a conserved region of the prefusion RSV F protein.
Nat Commun, 15, 2024
|
|
4MNA
 
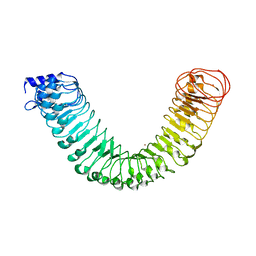 | | Crystal structure of the free FLS2 ectodomains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LRR receptor-like serine/threonine-protein kinase FLS2, ZINC ION | | Authors: | Chai, J, Han, Z, Sun, Y. | | Deposit date: | 2013-09-10 | | Release date: | 2013-12-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.998 Å) | | Cite: | Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex.
Science, 342, 2013
|
|
8XUO
 
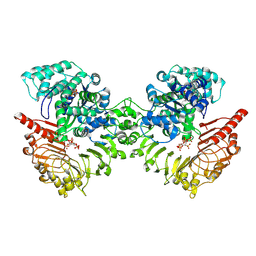 | | Cryo-EM structure of tomato NRC2 dimer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, NRC2 | | Authors: | Sun, Y, Ma, S.C, Chai, J.J. | | Deposit date: | 2024-01-13 | | Release date: | 2024-05-22 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Oligomerization-mediated autoinhibition and cofactor binding of a plant NLR.
Nature, 632, 2024
|
|
8XUQ
 
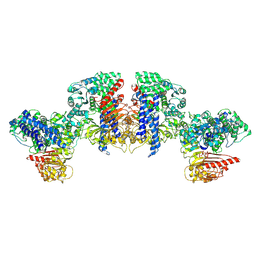 | | Cryo-EM structure of tomato NRC2 tetramer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, NRC2 | | Authors: | Sun, Y, Ma, S.C, Chai, J.J. | | Deposit date: | 2024-01-14 | | Release date: | 2024-05-22 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Oligomerization-mediated autoinhibition and cofactor binding of a plant NLR.
Nature, 632, 2024
|
|
8XUV
 
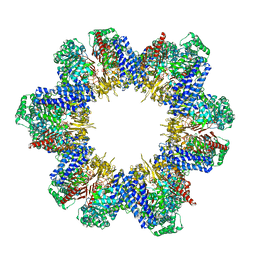 | | Cryo-EM structure of tomato NRC2 filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, NRC2 | | Authors: | Sun, Y, Ma, S.C, Chai, J.J. | | Deposit date: | 2024-01-14 | | Release date: | 2024-05-22 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Oligomerization-mediated autoinhibition and cofactor binding of a plant NLR.
Nature, 632, 2024
|
|
7BR0
 
 | | Crystal structure of AclR, a thioredoxin oxidoreductase fold protein carrying the CXXH catalytic motif | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Pyr_redox_2 domain-containing protein | | Authors: | Hara, K, Hashimoto, H, Maeda, N, Watanabe, K, Hertweck, C, Tsunematsu, Y. | | Deposit date: | 2020-03-26 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Specialized Flavoprotein Promotes Sulfur Migration and Spiroaminal Formation in Aspirochlorine Biosynthesis.
J.Am.Chem.Soc., 143, 2021
|
|
4KUL
 
 | | Crystal structure of N-terminal acetylated yeast Sir3 BAH domain V83P mutant | | Descriptor: | Regulatory protein SIR3 | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4KUI
 
 | | Crystal structure of N-terminal acetylated yeast Sir3 BAH domain | | Descriptor: | ACETIC ACID, ISOPROPYL ALCOHOL, Regulatory protein SIR3 | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4KUD
 
 | | Crystal structure of N-terminal acetylated Sir3 BAH domain D205N mutant in complex with yeast nucleosome core particle | | Descriptor: | Histone H2A.2, Histone H2B.1, Histone H3, ... | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.203 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
7BV5
 
 | | Crystal structure of the yeast heterodimeric ADAT2/3 | | Descriptor: | ZINC ION, tRNA-specific adenosine deaminase subunit TAD2, tRNA-specific adenosine deaminase subunit TAD3 | | Authors: | Xie, W, Liu, X, Chen, R, Sun, Y, Chen, R, Zhou, J, Tian, Q. | | Deposit date: | 2020-04-09 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the yeast heterodimeric ADAT2/3 deaminase.
Bmc Biol., 18, 2020
|
|
6SMH
 
 | | Cryo-electron microscopy structure of a RbcL-Raf1 supercomplex from Synechococcus elongatus PCC 7942 | | Descriptor: | Ribulose bisphosphate carboxylase large chain, Rubisco accumulation factor 1 (RAF1) peptide | | Authors: | Huang, F, Kong, W.-W, Sun, Y, Chen, T, Dykes, G.F, Jiang, Y.L, Liu, L.N. | | Deposit date: | 2019-08-21 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Rubisco accumulation factor 1 (Raf1) plays essential roles in mediating Rubisco assembly and carboxysome biogenesis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
