4WVC
 
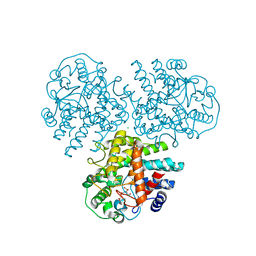 | | Crystal structure of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8 in complex with Tris and D-glycerate | | Descriptor: | (2R)-2,3-DIHYDROXYPROPANOIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Miyazaki, T, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-11-05 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and substrate-binding mode of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8.
J.Struct.Biol., 190, 2015
|
|
4WVA
 
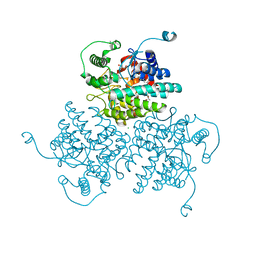 | | Crystal structure of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8 in complex with Tris | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Miyazaki, T, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-11-05 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure and substrate-binding mode of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8.
J.Struct.Biol., 190, 2015
|
|
4WVB
 
 | | Crystal structure of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8 in complex with glucose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Uncharacterized protein, ... | | Authors: | Miyazaki, T, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-11-05 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure and substrate-binding mode of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8.
J.Struct.Biol., 190, 2015
|
|
4XPO
 
 | | Crystal structure of a novel alpha-galactosidase from Pedobacter saltans | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-glucosidase | | Authors: | Miyazaki, T, Ishizaki, Y, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2015-01-17 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical characterization of novel bacterial alpha-galactosidases belonging to glycoside hydrolase family 31
Biochem.J., 469, 2015
|
|
4XPR
 
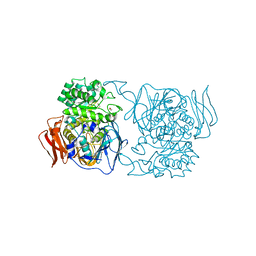 | | Crystal structure of the mutant D365A of Pedobacter saltans GH31 alpha-galactosidase | | Descriptor: | 1,2-ETHANEDIOL, Alpha-glucosidase | | Authors: | Miyazaki, T, Ishizaki, Y, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2015-01-17 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and biochemical characterization of novel bacterial alpha-galactosidases belonging to glycoside hydrolase family 31
Biochem.J., 469, 2015
|
|
4XPP
 
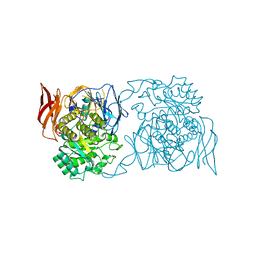 | | Crystal structure of Pedobacter saltans GH31 alpha-galactosidase complexed with D-galactose | | Descriptor: | 1,2-ETHANEDIOL, Alpha-glucosidase, beta-D-galactopyranose | | Authors: | Miyazaki, T, Ishizaki, Y, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2015-01-17 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical characterization of novel bacterial alpha-galactosidases belonging to glycoside hydrolase family 31
Biochem.J., 469, 2015
|
|
4XPQ
 
 | | Crystal structure of Pedobacter saltans GH31 alpha-galactosidase complexed with L-fucose | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-glucosidase, ... | | Authors: | Miyazaki, T, Ishizaki, Y, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2015-01-17 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and biochemical characterization of novel bacterial alpha-galactosidases belonging to glycoside hydrolase family 31
Biochem.J., 469, 2015
|
|
4XPS
 
 | | Crystal structure of the mutant D365A of Pedobacter saltans GH31 alpha-galactosidase complexed with p-nitrophenyl-alpha-galactopyranoside | | Descriptor: | 1,2-ETHANEDIOL, Alpha-glucosidase, P-NITROPHENOL, ... | | Authors: | Miyazaki, T, Ishizaki, Y, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2015-01-17 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical characterization of novel bacterial alpha-galactosidases belonging to glycoside hydrolase family 31
Biochem.J., 469, 2015
|
|
5GW7
 
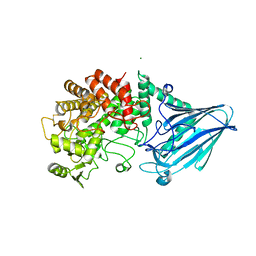 | |
7BOO
 
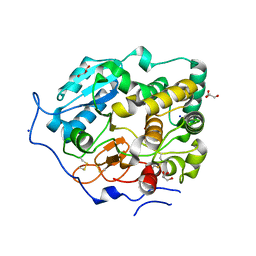 | | Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, apo form | | Descriptor: | Alpha-1,2-mannosyltransferase (Ktr4), putative, GLYCEROL, ... | | Authors: | Hira, D, Onoue, T, Oka, T. | | Deposit date: | 2020-03-19 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the core-mannan biosynthesis of cell wall fungal-type galactomannan in Aspergillus fumigatus .
J.Biol.Chem., 295, 2020
|
|
7BOP
 
 | | Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, Mn/GDP-form | | Descriptor: | Alpha-1,2-mannosyltransferase (Ktr4), putative, GLYCEROL, ... | | Authors: | Hira, D, Onoue, T, Oka, T. | | Deposit date: | 2020-03-19 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the core-mannan biosynthesis of cell wall fungal-type galactomannan in Aspergillus fumigatus .
J.Biol.Chem., 295, 2020
|
|
7YH7
 
 | | SARS-CoV-2 spike in complex with neutralizing antibody NIV-8 (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-07-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
7YH6
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, NIV-8 Fab light chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-07-12 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
4EN6
 
 | | Crystal structure of HA70 (HA3) subcomponent of Clostridium botulinum type C progenitor toxin in complex with alpha 2-3-sialyllactose | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Hemagglutinin components HA-22/23/53, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Yamashita, S, Yoshida, H, Tonozuka, T, Nishikawa, A, Kamitori, S. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Carbohydrate recognition mechanism of HA70 from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides
Febs Lett., 586, 2012
|
|
4EN7
 
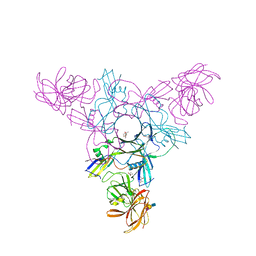 | | Crystal structure of HA70 (HA3) subcomponent of Clostridium botulinum type C progenitor toxin in complex with alpha 2-3-sialyllactosamine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Hemagglutinin components HA-22/23/53, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yamashita, S, Yoshida, H, Tonozuka, T, Nishikawa, A, Kamitori, S. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Carbohydrate recognition mechanism of HA70 from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides
Febs Lett., 586, 2012
|
|
4EN8
 
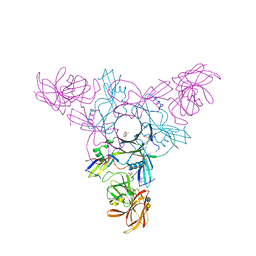 | | Crystal structure of HA70 (HA3) subcomponent of Clostridium botulinum type C progenitor toxin in complex with alpha 2-6-sialyllactose | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Hemagglutinin components HA-22/23/53, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Yamashita, S, Yoshida, H, Tonozuka, T, Nishikawa, A, Kamitori, S. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Carbohydrate recognition mechanism of HA70 from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides
Febs Lett., 586, 2012
|
|
4EN9
 
 | | Crystal structure of HA70 (HA3) subcomponent of Clostridium botulinum type C progenitor toxin in complex with alpha 2-6-sialyllactosamine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Hemagglutinin components HA-22/23/53, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yamashita, S, Yoshida, H, Tonozuka, T, Nishikawa, A, Kamitori, S. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Carbohydrate recognition mechanism of HA70 from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides
Febs Lett., 586, 2012
|
|
1BVZ
 
 | | ALPHA-AMYLASE II (TVAII) FROM THERMOACTINOMYCES VULGARIS R-47 | | Descriptor: | PROTEIN (ALPHA-AMYLASE II) | | Authors: | Kamitori, S, Kondo, S, Okuyama, K, Yokota, T, Shimura, Y, Tonozuka, T, Sakano, Y. | | Deposit date: | 1998-09-22 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Thermoactinomyces vulgaris R-47 alpha-amylase II (TVAII) hydrolyzing cyclodextrins and pullulan at 2.6 A resolution.
J.Mol.Biol., 287, 1999
|
|
5CA3
 
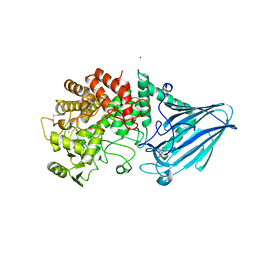 | |
1V94
 
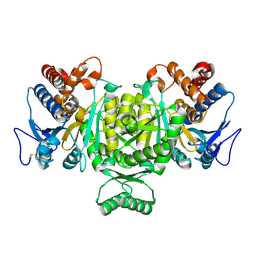 | | Crystal structure of isocitrate dehydrogenase from Aeropyrum pernix | | Descriptor: | isocitrate dehydrogenase | | Authors: | Jeong, J.-J, Sonoda, T, Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2004-01-20 | | Release date: | 2005-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of isocitrate dehydrogenase from Aeropyrum pernix
Proteins, 55, 2004
|
|
5XH9
 
 | | Aspergillus kawachii beta-fructofuranosidase | | Descriptor: | Extracellular invertase, SODIUM ION | | Authors: | Nagaya, M, Tonozuka, T. | | Deposit date: | 2017-04-19 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a beta-fructofuranosidase with high transfructosylation activity from Aspergillus kawachii
Biosci. Biotechnol. Biochem., 81, 2017
|
|
5XHA
 
 | |
5XH8
 
 | |
3AH4
 
 | | HA1 subcomponent of botulinum type C progenitor toxin complexed with galactose | | Descriptor: | Main hemagglutinin component, beta-D-galactopyranose | | Authors: | Nakamura, T, Tonozuka, T, Ide, A, Yuzawa, T, Oguma, K, Nishikawa, A. | | Deposit date: | 2010-04-13 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Sugar-binding sites of the HA1 subcomponent of Clostridium botulinum type C progenitor toxin
J.Mol.Biol., 376, 2008
|
|
3AH2
 
 | | HA1 subcomponent of botulinum type C progenitor toxin complexed with N-acetylgalactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, Main hemagglutinin component | | Authors: | Nakamura, T, Tonozuka, T, Ide, A, Yuzawa, T, Oguma, K, Nishikawa, A. | | Deposit date: | 2010-04-13 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Sugar-binding sites of the HA1 subcomponent of Clostridium botulinum type C progenitor toxin
J.Mol.Biol., 376, 2008
|
|
