1WRJ
 
 | |
4IJ0
 
 | | Structures of DNA duplexes containing O6-carboxymethylguanine, a lesion associated with gastrointestinal cancer, reveal a mechanism for inducing transition mutation | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*(C6G)P*AP*AP*TP*TP*CP*GP*CP*G)-3'), STRONTIUM ION | | Authors: | Zhang, F, Suzuki, K, Tsunoda, M, Wilkinson, O, Millington, C.L, Williams, D.M, Morishita, E.C, Takenaka, A. | | Deposit date: | 2012-12-20 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structures of DNA duplexes containing O6-carboxymethylguanine, a lesion associated with gastrointestinal cancer, reveal a mechanism for inducing pyrimidine transition mutations
Nucleic Acids Res., 41, 2013
|
|
4ITD
 
 | | Structures of DNA duplexes containing O6-carboxymethylguanine, a lesion associated with gastrointestinal cancer, reveal a mechanism for inducing transition mutation | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*(C6G)P*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Zhang, F, Suzuki, K, Tsunoda, M, Wilkinson, O, Millington, C.L, Williams, D.M, Morishita, E.C, Takenaka, A. | | Deposit date: | 2013-01-18 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures of DNA duplexes containing O6-carboxymethylguanine, a lesion associated with gastrointestinal cancer, reveal a mechanism for inducing pyrimidine transition mutations
Nucleic Acids Res., 41, 2013
|
|
1J8L
 
 | | Molecular and Crystal Structure of D(CGCAAATTMO4CGCG): the Watson-Crick Type N4-Methoxycytidine/Adenosine Base Pair in B-DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*(C45)P*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Hossain, M.T, Sunami, T, Tsunoda, M, Hikima, T, Chatake, T, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 2001-05-22 | | Release date: | 2001-09-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic studies on damaged DNAs IV. N(4)-methoxycytosine shows a second face for Watson-Crick base-pairing, leading to purine transition mutagenesis.
Nucleic Acids Res., 29, 2001
|
|
1I47
 
 | | MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGAATT(MO4)CGCG): THE WATSON-CRICK TYPE AND WOBBLE N4-METHOXYCYTIDINE/GUANOSINE BASE PAIRS IN B-DNA | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(C45)P*GP*CP*GP)-3', MAGNESIUM ION | | Authors: | Hossain, M.T, Hikima, T, Chatake, T, Tsunoda, M, Sunami, T, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 2001-02-20 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic studies on damaged DNAs: III. N(4)-methoxycytosine can form both Watson-Crick type and wobbled base pairs in a B-form duplex
J.Biochem.(Tokyo), 130, 2001
|
|
1V53
 
 | | The crystal structure of 3-isopropylmalate dehydrogenase from Bacillus coagulans | | Descriptor: | 3-isopropylmalate dehydrogenase | | Authors: | Fujita, K, Minami, H, Suzuki, K, Tsunoda, M, Sekiguchi, T, Mizui, R, Tsuzaki, S, Nakamura, S, Takenaka, A. | | Deposit date: | 2003-11-20 | | Release date: | 2005-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The crystal structure of 3-isopropylmalate dehydrogenase from Bacillus coagulans
To be Published
|
|
1V5B
 
 | | The Structure Of The Mutant, S225A and E251L, Of 3-Isopropylmalate Dehydrogenase From Bacillus Coagulans | | Descriptor: | 3-isopropylmalate dehydrogenase, SULFATE ION | | Authors: | Fujita, K, Minami, H, Suzuki, K, Tsunoda, M, Sekiguchi, T, Mizui, R, Tsuzaki, S, Nakamura, S, Takenaka, A. | | Deposit date: | 2003-11-22 | | Release date: | 2005-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of a highly thermo-stabilized mutant of 3-isopropylmalate dehydrogenase from Bacillus coagulans: An evaluation of local packing density in the hydrophobic core
To be Published
|
|
1V59
 
 | | Crystal structure of yeast lipoamide dehydrogenase complexed with NAD+ | | Descriptor: | Dihydrolipoamide dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Adachi, W, Suzuki, K, Tsunoda, M, Sekiguchi, T, Reed, L.J, Takenaka, A. | | Deposit date: | 2003-11-21 | | Release date: | 2005-02-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of yeast lipoamide dehydrogenase complexed with NAD+
To be Published
|
|
3A31
 
 | | Crystal structure of putative threonyl-tRNA synthetase ThrRS-1 from Aeropyrum pernix (selenomethionine derivative) | | Descriptor: | Probable threonyl-tRNA synthetase 1, SULFATE ION, ZINC ION | | Authors: | Shimizu, S, Juan, E.C.M, Miyashita, Y, Sato, Y, Hoque, M.M, Suzuki, K, Yogiashi, M, Tsunoda, M, Dock-Bregeon, A.-C, Moras, D, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2009-06-07 | | Release date: | 2009-10-27 | | Last modified: | 2013-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two complementary enzymes for threonylation of tRNA in crenarchaeota: crystal structure of Aeropyrum pernix threonyl-tRNA synthetase lacking a cis-editing domain
J.Mol.Biol., 394, 2009
|
|
3A32
 
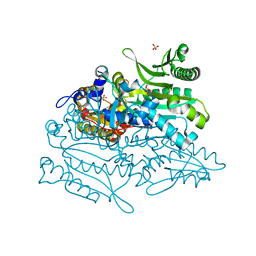 | | Crystal structure of putative threonyl-tRNA synthetase ThrRS-1 from Aeropyrum pernix | | Descriptor: | Probable threonyl-tRNA synthetase 1, SULFATE ION, ZINC ION | | Authors: | Shimizu, S, Juan, E.C.M, Miyashita, Y, Sato, Y, Hoque, M.M, Suzuki, K, Yogiashi, M, Tsunoda, M, Dock-Bregeon, A.-C, Moras, D, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2009-06-07 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Two complementary enzymes for threonylation of tRNA in crenarchaeota: crystal structure of Aeropyrum pernix threonyl-tRNA synthetase lacking a cis-editing domain
J.Mol.Biol., 394, 2009
|
|
3VDR
 
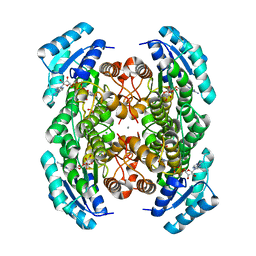 | | Crystal structure of D-3-hydroxybutyrate dehydrogenase, prepared in the presence of the substrate D-3-hydroxybutyrate and NAD(+) | | Descriptor: | (3R)-3-hydroxybutanoic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ACETOACETIC ACID, ... | | Authors: | Hoque, M.M, Shimizu, S, Juan, E.C.M, Sato, Y, Hossain, M.T, Yamamoto, T, Imamura, S, Amano, H, Suzuki, K, Sekiguchi, T, Tsunoda, M, Takenaka, A. | | Deposit date: | 2012-01-06 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of D-3-hydroxybutyrate dehydrogenase prepared in the presence of the substrate D-3-hydroxybutyrate and NAD+.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3W8D
 
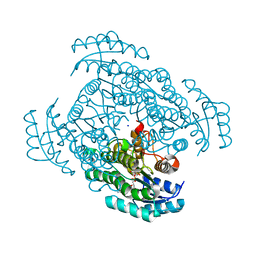 | | Crystal structure of D-3-hydroxybutyrate dehydrogenase from Alcaligenes faecalis complexed with NAD+ and an inhibitor methylmalonate | | Descriptor: | CHLORIDE ION, D-3-hydroxybutyrate dehydrogenase, METHYLMALONIC ACID, ... | | Authors: | Kanazawa, H, Tsunoda, M, Hoque, M.M, Suzuki, K, Yamamoto, T, Takenaka, A. | | Deposit date: | 2013-03-12 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | X-ray diffraction of D-3-hydroxybutyrate dehydrogenase from Alcaligenes faecalis complexed with NAD+ and methylmalonate
To be Published
|
|
3W8E
 
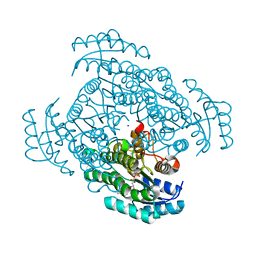 | | Crystal structure of D-3-hydroxybutyrate dehydrogenase from Alcaligenes faecalis complexed with NAD+ and a substrate D-3-hydroxybutyrate | | Descriptor: | (3R)-3-hydroxybutanoic acid, D-3-hydroxybutyrate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Kanazawa, H, Tsunoda, M, Hoque, M.M, Suzuki, K, Yamamoto, T, Takenaka, A. | | Deposit date: | 2013-03-12 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | High resolution X-ray diffraction of D-3-hydroxybutyrate dehydrogenase from Alcaligenes faecalis complexed with NAD+ and D-3-hydroxybutyrate
To be Published
|
|
3W8F
 
 | | Crystal structure of D-3-hydroxybutyrate dehydrogenase from Alcaligenes faecalis complexed with NAD+ and an inhibitor malonate | | Descriptor: | CHLORIDE ION, D-3-hydroxybutyrate dehydrogenase, MALONIC ACID, ... | | Authors: | Kanazawa, H, Tsunoda, M, Hoque, M.M, Suzuki, K, Yamamoto, T, Takenaka, A. | | Deposit date: | 2013-03-12 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | X-ray diffraction of D-3-hydroxybutyrate dehydrogenase from Alcaligenes faecalis complexed with NAD+ and malonate
To be Published
|
|
6J6U
 
 | | Rat PTPRZ D1-D2 domain | | Descriptor: | Receptor-type tyrosine-protein phosphatase zeta | | Authors: | Sugawara, H. | | Deposit date: | 2019-01-15 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | A head-to-toe dimerization has physiological relevance for ligand-induced inactivation of protein tyrosine receptor type Z.
J.Biol.Chem., 294, 2019
|
|
5H08
 
 | | Human PTPRZ D1 domain complexed with NAZ2329 | | Descriptor: | 3-{[2-Ethoxy-5-(trifluoromethyl)benzyl]sulfanyl}-N-(phenylsulfonyl)thiophene-2-carboxamide, Receptor-type tyrosine-protein phosphatase zeta | | Authors: | Sugawara, H. | | Deposit date: | 2016-10-04 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Targeting PTPRZ inhibits stem cell-like properties and tumorigenicity in glioblastoma cells
Sci Rep, 7, 2017
|
|
5X7Y
 
 | | Crystal Structure of the Dog Lipocalin Allergen Can f 6 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Lipocalin-Can f 6 allergen | | Authors: | Yamamoto, K, Otani, T, Sugiura, K, Nakatsuji, M, Nishimura, S, Inui, T. | | Deposit date: | 2017-02-28 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the dog allergen Can f 6 and structure-based implications of its cross-reactivity with the cat allergen Fel d 4.
Sci Rep, 9, 2019
|
|
5AWX
 
 | | Crystal structure of Human PTPRZ D1 domain | | Descriptor: | BROMIDE ION, Receptor-type tyrosine-protein phosphatase zeta | | Authors: | Sugawara, H. | | Deposit date: | 2015-07-10 | | Release date: | 2016-02-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Small-molecule inhibition of PTPRZ reduces tumor growth in a rat model of glioblastoma
Sci Rep, 6, 2016
|
|
6LE4
 
 | |
6LDO
 
 | |
5B1I
 
 | |
7JRI
 
 | |
7MPB
 
 | | SARS Coronavirus-2 Main Protease 3CL-pro binding Ascorbate | | Descriptor: | 3C-like proteinase, ASCORBIC ACID, TRIFLUOROETHANOL | | Authors: | Pandey, S, Malla, T.N, Stojkovic, E.A, Schmidt, M. | | Deposit date: | 2021-05-04 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Vitamin C inhibits SARS coronavirus-2 main protease essential for viral replication
Biorxiv, 2021
|
|
6PU2
 
 | | Dark, Mutant H275T , 100K, PCM Myxobacterial Phytochrome, P2 | | Descriptor: | 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Pandey, S, Schmidt, M, Stojkovic, E.A. | | Deposit date: | 2019-07-16 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-resolution crystal structures of a myxobacterial phytochrome at cryo and room temperatures.
Struct Dyn., 6, 2019
|
|
6PTQ
 
 | | Dark, Room Temperature, PCM Myxobacterial Phytochrome, P2, Wild Type | | Descriptor: | 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, BENZAMIDINE, Photoreceptor-histidine kinase BphP | | Authors: | Pandey, S, Schmidt, M, Stojkovic, E.A. | | Deposit date: | 2019-07-16 | | Release date: | 2019-10-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution crystal structures of a myxobacterial phytochrome at cryo and room temperatures.
Struct Dyn., 6, 2019
|
|
