4GCW
 
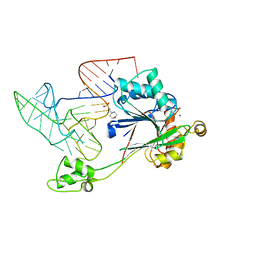 | | Crystal structure of RNase Z in complex with precursor tRNA(Thr) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ribonuclease Z, TRNA(THR), ... | | Authors: | Pellegrini, O, Li de la Sierra-Gallay, I, Piton, J, Gilet, L, Condon, C. | | Deposit date: | 2012-07-31 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Activation of tRNA Maturation by Downstream Uracil Residues in B. subtilis
Structure, 20, 2012
|
|
4JZU
 
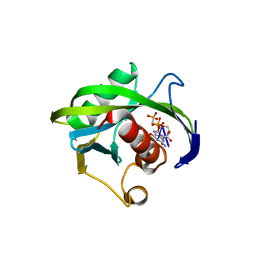 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH bound to a non-hydrolysable triphosphorylated dinucleotide RNA (pcp-pGpG) - first guanosine residue in guanosine binding pocket | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, RNA (5'-R(*(GCP)P*G)-3'), RNA PYROPHOSPHOHYDROLASE | | Authors: | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | Deposit date: | 2013-04-03 | | Release date: | 2013-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JZT
 
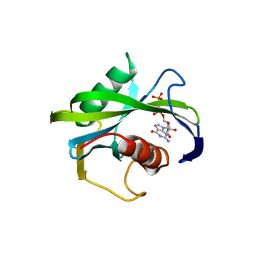 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH (E68A mutant) bound to GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, dGTP pyrophosphohydrolase | | Authors: | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | Deposit date: | 2013-04-03 | | Release date: | 2013-05-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JZV
 
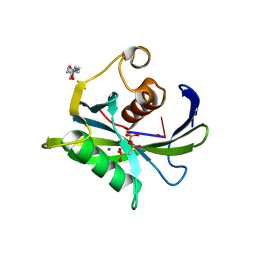 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH bound to a non-hydrolysable triphosphorylated dinucleotide RNA (pcp-pGpG) - second guanosine residue in guanosine binding pocket | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, RNA (5'-R(*(GCP)P*G)-3'), ... | | Authors: | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | Deposit date: | 2013-04-03 | | Release date: | 2013-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6RCY
 
 | |
6EHI
 
 | | NucT from Helicobacter pylori | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Celma, L, Li de la Sierra-Gallay, I, Quevillon-Cheruel, S. | | Deposit date: | 2017-09-13 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural basis for the substrate selectivity of Helicobacter pylori NucT nuclease activity.
PLoS ONE, 12, 2017
|
|
5IKP
 
 | | Crystal structure of human brain glycogen phosphorylase bound to AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glycogen phosphorylase, brain form, ... | | Authors: | Mathieu, C, Li de la Sierra-Gallay, I, Xu, X, Haouz, A, Rodrigues-Lima, F. | | Deposit date: | 2016-03-03 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Insights into Brain Glycogen Metabolism: THE STRUCTURE OF HUMAN BRAIN GLYCOGEN PHOSPHORYLASE.
J.Biol.Chem., 291, 2016
|
|
5IKO
 
 | | Crystal structure of human brain glycogen phosphorylase | | Descriptor: | Glycogen phosphorylase, brain form, HEXAETHYLENE GLYCOL, ... | | Authors: | Mathieu, C, Li de la Sierra-Gallay, I, Xu, X, Haouz, A, Rodrigues-Lima, F. | | Deposit date: | 2016-03-03 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into Brain Glycogen Metabolism: THE STRUCTURE OF HUMAN BRAIN GLYCOGEN PHOSPHORYLASE.
J.Biol.Chem., 291, 2016
|
|
6S84
 
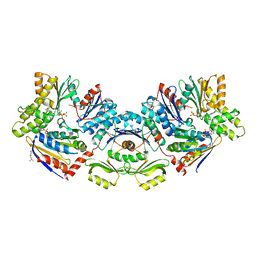 | | TsaBDE complex from Thermotoga maritima | | Descriptor: | ATPase YjeE, predicted to have essential role in cell wall biosynthesis, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Missoury, S, Li-de-La-Sierra-Gallay, I, van Tilbeurgh, H. | | Deposit date: | 2019-07-08 | | Release date: | 2019-07-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | The structure of the TsaB/TsaD/TsaE complex reveals an unexpected mechanism for the bacterial t6A tRNA-modification.
Nucleic Acids Res., 46, 2018
|
|
4XL5
 
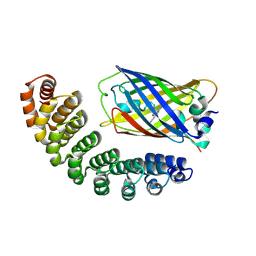 | | X-ray structure of bGFP-A / EGFP complex | | Descriptor: | Green fluorescent protein, bGFP-A | | Authors: | Chevrel, A, Urvoas, A, Li de la Sierra-Gallay, I, Van Tilbeurgh, H, Minard, P, Valerio-Lepiniec, M. | | Deposit date: | 2015-01-13 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specific GFP-binding artificial proteins ( alpha Rep): a new tool for in vitro to live cell applications.
Biosci.Rep., 35, 2015
|
|
4XVP
 
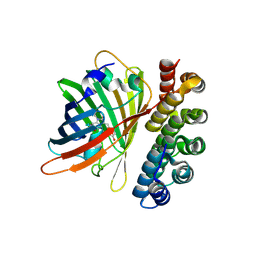 | | X-ray structure of bGFP-C / EGFP complex | | Descriptor: | BGFP-C, Green fluorescent protein | | Authors: | Chevrel, A, Urvoas, A, Li de la Sierra-Gallay, I, Van Tilbeurgh, H, Minard, P, Valerio-Lepiniec, M. | | Deposit date: | 2015-01-27 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Specific GFP-binding artificial proteins ( alpha Rep): a new tool for in vitro to live cell applications.
Biosci.Rep., 35, 2015
|
|
4H18
 
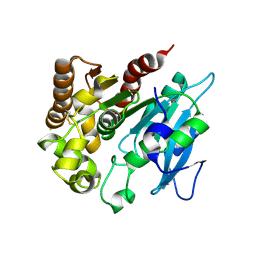 | | Three dimensional structure of corynomycoloyl tranferase C | | Descriptor: | Cmt1, MAGNESIUM ION | | Authors: | Huc, E, de Sousa D'Auria, C, Li de la Sierra-Gallay, I, Salmeron, C.H, van Tilbeurgh, H, Bayan, N, Houssin, C.H, Daffe, M, Tropis, M. | | Deposit date: | 2012-09-10 | | Release date: | 2013-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Identification of a mycoloyl transferase selectively involved in o-acylation of polypeptides in corynebacteriales.
J.Bacteriol., 195, 2013
|
|
6QER
 
 | |
6G4J
 
 | |
5DBK
 
 | |
6HU8
 
 | |
6HUA
 
 | | the competence regulator ComR from Streptococcus vestibularis in complex with its cognate signaling peptide XIP | | Descriptor: | Uncharacterized protein, XIP signaling peptide | | Authors: | Nessler, S, Thuillier, J, Ledesma, L, Hols, P. | | Deposit date: | 2018-10-05 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.389 Å) | | Cite: | Molecular dissection of pheromone selectivity in the competence signaling system ComRS of streptococci.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6NAK
 
 | | BACTERIAL PROTEIN COMPLEX TM BDE complex | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, TsaE, ... | | Authors: | Stec, B. | | Deposit date: | 2018-12-05 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Discovery of the Universal tRNA Binding Mode for the TsaD-like Components of the t6A tRNA Modification Pathway
Biophysica, 3, 2023
|
|
8A3V
 
 | | Crystal structure of the Vibrio cholerae replicative helicase (VcDnaB) in complex with its loader protein (VcDciA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DUF721 domain-containing protein, MAGNESIUM ION, ... | | Authors: | Walbott, H, Quevillon-Cheruel, S, Cargemel, C. | | Deposit date: | 2022-06-09 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The LH-DH module of bacterial replicative helicases is the common binding site for DciA and other helicase loaders.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
3UQZ
 
 | |
