7K3T
 
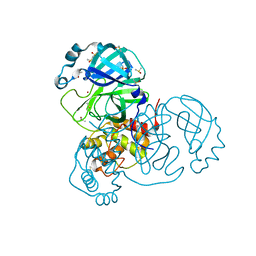 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) at 1.2 A Resolution and a Possible Capture of Zinc Binding Intermediate | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Andi, B, Kumaran, D, Kreitler, D.F, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-09-13 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
9C5S
 
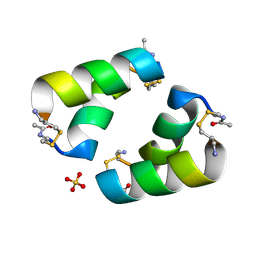 | | Disulfide-linked, antiparallel p53-derived peptide dimer (CV1) | | Descriptor: | Cellular tumor antigen p53, SULFATE ION | | Authors: | Vithanage, N, Kreitler, D.K, DiGiorno, M.C, Victorio, C.G, Sawyer, N, Outlaw, V.K. | | Deposit date: | 2024-06-06 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Structural Characterization of Disulfide-Linked p53-Derived Peptide Dimers.
Res Sq, 2024
|
|
4GWT
 
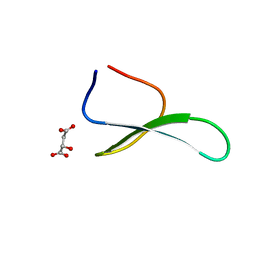 | | Structure of racemic Pin1 WW domain cocrystallized with DL-malic acid | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Mortenson, D.E, Yun, H.G, Gellman, S.H, Forest, K.T. | | Deposit date: | 2012-09-03 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Evidence for small-molecule-mediated loop stabilization in the structure of the isolated Pin1 WW domain.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8GDW
 
 | |
8GF4
 
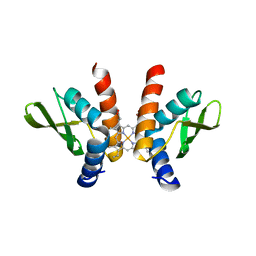 | |
8GBK
 
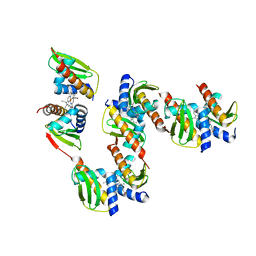 | |
4GWV
 
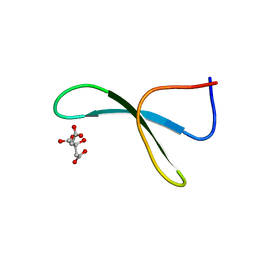 | | Structure of racemic Pin1 WW domain cocrystallized with tri-ammonium citrate | | Descriptor: | CITRATE ANION, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Mortenson, D.E, Yun, H.G, Gellman, S.H, Forest, K.T. | | Deposit date: | 2012-09-03 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Evidence for small-molecule-mediated loop stabilization in the structure of the isolated Pin1 WW domain.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7U1B
 
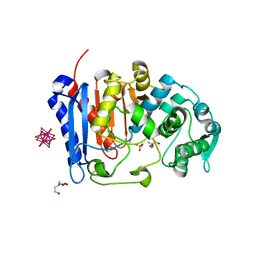 | | Crystal structure of EstG in complex with tantalum cluster | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BICINE, Beta-lactamase domain-containing protein, ... | | Authors: | Chen, Z, Gabelli, S.B. | | Deposit date: | 2022-02-20 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | EstG is a novel esterase required for cell envelope integrity in Caulobacter.
Curr.Biol., 33, 2023
|
|
7U1C
 
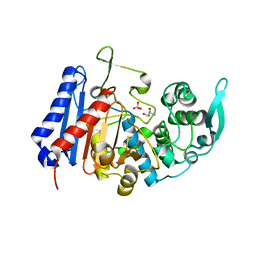 | | Structure of EstG crystalized with SO4 and Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase domain-containing protein, SODIUM ION, ... | | Authors: | Gabelli, S.B, Chen, Z. | | Deposit date: | 2022-02-20 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | EstG is a novel esterase required for cell envelope integrity in Caulobacter.
Curr.Biol., 33, 2023
|
|
7UDA
 
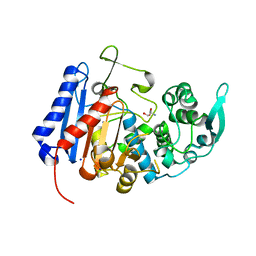 | | Structure of the EstG | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase domain-containing protein, SODIUM ION | | Authors: | Chen, Z, Gabelli, S.B. | | Deposit date: | 2022-03-18 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | EstG is a novel esterase required for cell envelope integrity in Caulobacter.
Curr.Biol., 33, 2023
|
|
8FM6
 
 | |
6PRL
 
 | |
6PYQ
 
 | |
6PZ6
 
 | |
6O4M
 
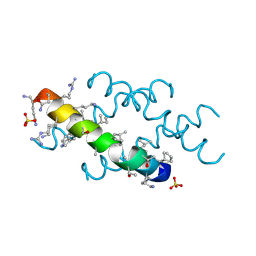 | | Racemic melittin | | Descriptor: | D-Melittin, Melittin, SULFATE ION | | Authors: | Kurgan, K.W, Bingman, C.A, Gellman, S.H, Forest, K.T. | | Deposit date: | 2019-02-28 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Retention of Native Quaternary Structure in Racemic Melittin Crystals.
J.Am.Chem.Soc., 141, 2019
|
|
6V3V
 
 | |
6VAS
 
 | |
8G83
 
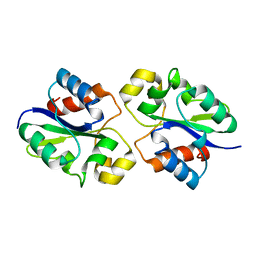 | | Structure of NAD+ consuming protein Acinetobacter baumannii TIR domain | | Descriptor: | NAD(+) hydrolase AbTIR | | Authors: | Klontz, E.H, Wang, Y, Glendening, G, Carr, J, Tsibouris, T, Buddula, S, Nallar, S, Soares, A, Snyder, G.A. | | Deposit date: | 2023-02-17 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | The structure of NAD + consuming protein Acinetobacter baumannii TIR domain shows unique kinetics and conformations.
J.Biol.Chem., 299, 2023
|
|
